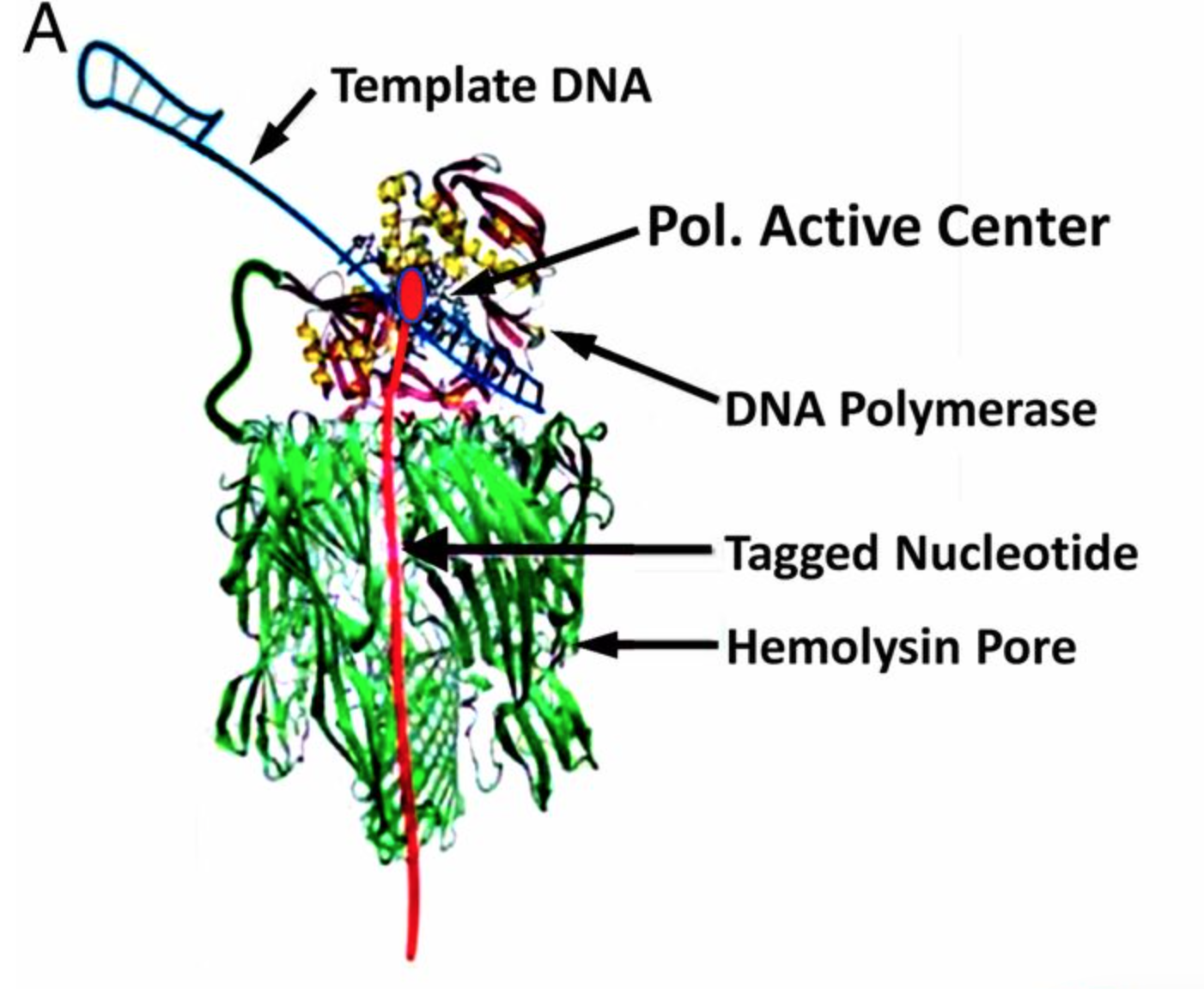
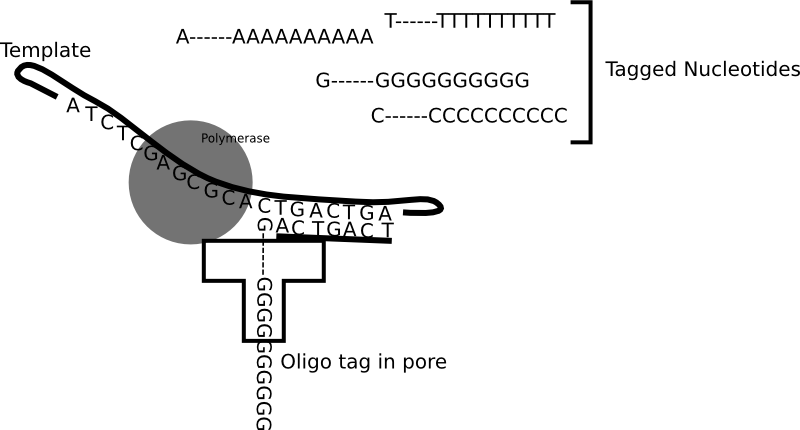
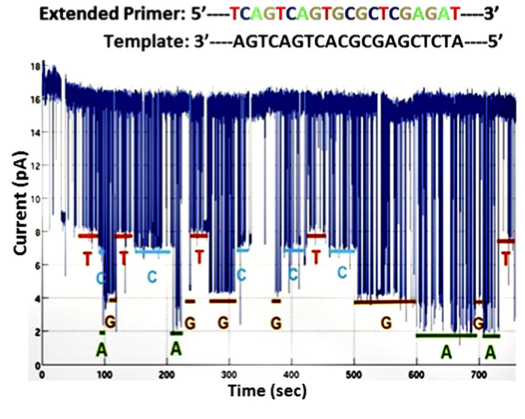
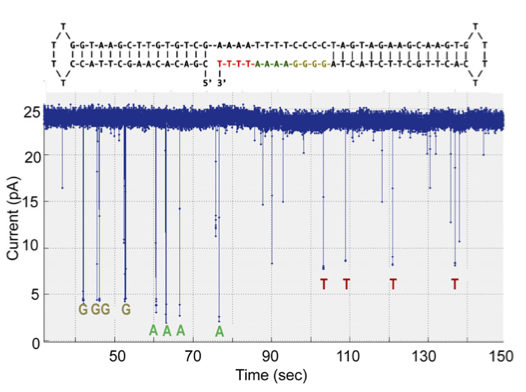
In my previous post on the new Genia paper, I mentioned the following quote from their paper: “The applied voltage is adjusted to ensure that, in a majority of cases, one and only one pore is inserted into the membranes of each well.”. It’s a one line note, but it intrigued me. First, a little background on why this is a moderately interesting development.
In general protein nanopores insert themselves into bilayers. No applied voltage or particularly special conditions are required for this, it is after all the purpose for which they have evolved. There is of course nothing to stop them from inserting multiple times into the same bilayer, and for sequencing that causes issues. The cartoon below show the signal from an ideal single nanopore:
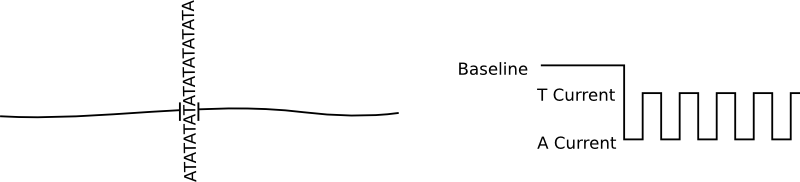
Great! A nice clean signal. But if we have two pores inserted, as well as a shifted baseline we get a signal that much harder to interpret and can not be basecalled:
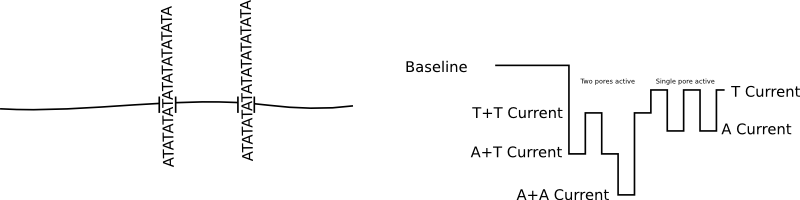
Nanopore based sequencers have in general addressed this by throwing channels at the problem and assuming that you’ll only get 37% of the wells occupied by a single pore (poisson limited). Which is why you see statements like:
The MinION uses a chip that contains ∼2000 robust polymer, rather than lipid bilayer, membranes over microwells that are individually electrically addressed. Assuming a Poisson distribution, a maximum of 37% of the membranes will contain a single nanopore when pores are allowed to insert randomly at the most favorable concentration. [From this paper]
This loses 63% of the system throughput and wastes a significant portion of the chip real-estate. Genia appear to have an at least partial solution to this problem.
The technique is described in a patent (US20140203464 A1) and is really quite simple. I’d guess as with most things the devil is in the details. What they do is take advantage of electroporation, electroporation is general lab technqiue used to bash holes in cell bilayers to introduce DNA or other chemicals into a cell. From what I’ve read it usually uses 100s of volts.
Genia appear to be using a voltage in the range of 200 to 700mV to weaken the bilayer and promote nanopore insertion. The assumption is (and again, data is somewhat limited) that this relatively large voltage increases the chance of a pore inserting [1].
The pore is I guess constantly attempting to insert into the bilayer with some low probability. The increased bias pushes this probability up significantly. Because Genia have designed their chips with independent control of the biases voltages on each well they can ramp up the bias voltage, check for insertion and if a single pore has inserted reduce the bias and begin detection. They use a relatively low concentration of pores, so that the difference between the probability of inserting in a normal well and one with a high bias voltage is significant.
This seems like a neat approach, and if it really works is quite a nice hack to get round the poisson limit. It’s a technique that should be generally applicable to protein nanopore systems (I assumed they’d need to license from Genia, if the patent holds up).
Disclosure/Disclaimer: I have worked and continue to work in the DNA sequencing industry. I own stock in DNA sequencing companies. While I have tried to be unbiased this represents my opinion and speculation only. I recommend reading the publicly available paper for yourself.
[1] 200 to 700mV is significantly higher than the bias generally use for base detection (which is in the 100 to 200mV range).
Relevant section of the patent
For reference, the relevant section of the patent is below:
[0071] In contrast to the self-assembly techniques, the present application discloses an electroporation technique in which an agitation electrical stimulus is applied across the lipid bilayer membrane to disrupt the lipid bilayer and initiate the formation of a nanopore in the lipid bilayer. The concentration of the nanopore forming solution may be readily maintained at a sufficiently low enough level such that no nanopores are inserted automatically into the lipid bilayers. In addition, the agitation electrical stimulus may be applied in such a way that only a single nanopore is inserted into each lipid bilayer.
[0072] Electroporation allows lower amounts (lower concentration) of hemolysin protein in the nanopore forming solution to achieve pore insertions. In some embodiments, the concentration of pore protein is at a level such that when applied to an array of bilayer-covered sensors, less than 10 percent of the bilayers have pores inserted in an uncontrolled manner. Using a lower concentration of pore protein has a number of advantages. For example, a user can have better control of the pore insertion because once a pore is inserted by electroporation, a second pore is unlikely to be inserted by itself. Using a lower concentration of pore protein is more economical. In addition, a lower concentration of pore protein is safer for the user to handle. For example, a high concentration of pore protein spilled on human skin may cause a severe rash, thus posing a health hazard.
[0073] If the electroporation technique described herein is not used, but a solution of protein pores is simply applied to an array of independent bilayers, it is well known to those skilled in the art that the insertion of protein pores into lipid bilayers will follow a Poisson distribution. This independent, randomness in pore insertion over time means that not only pore insertions are uncontrolled but that insertions are independent of prior events and therefore illustrates that more than one pore can be inserted into a single bilayer. Such a random system limits the percentage of single pore instances that one may obtain using concentration of pore protein alone. Changing a nanopore system from a random set of events with a Poisson distribution to a system with active, deterministic pore insertion (not following the Poisson distribution) gives the new system great advantage in attaining a higher percentage of bilayers with single pores.
[0074] The level of the agitation electrical stimulus at which a nanopore is inserted into a lipid bilayer may vary based on various factors, including salt concentration, temperature, the type of lipid membrane material, size of the lipid bilayer, and the like. In some embodiments, about 80-90% of the nanopores are inserted when the magnitude of the agitation electrical stimulus is within the range of 200 mV to 450 mV. The rest of the nanopores may be inserted at lower voltage levels, e.g., 100 mV or at higher levels, e.g., 700 mV.
[0075] In some embodiments, the steps of applying the agitation electrical stimulus, applying the negative stimulus (reverse oxidation stimulus), and applying the measuring electrical stimulus for detecting whether a nanopore has been properly formed are iterated in a loop, until a nanopore is finally formed in the lipid bilayer. Initially, the magnitude of the agitation electrical stimulus is set at a lower level. The magnitude of the agitation electrical stimulus is then gradually increased after each iteration. For example, the initial agitation electrical stimulus may be set at a lower level, such as 60 mV. After each iteration, if a nanopore is still not properly inserted, then the agitation electrical stimulus is adjusted by a small increment, e.g., 2 mV. This process may be repeated until a nanopore is finally inserted into the lipid bilayer, or until the lipid bilayer is damaged or erased by the agitation electrical stimulus, or until the agitation electrical stimulus has reached a predetermined maximum threshold, e.g., 700 mV. By gradually increasing the agitation electrical stimulus level (e.g., from 60 mV to 700 mV) as describe above, a higher percentage of the cells of the nanopore chip will have one nanopore properly inserted into a lipid bilayer.
[0076] In FIG. 5A, agitation electrical stimulus is shown as a positive voltage. However, a negative agitation electrical stimulus may be used in some embodiments as well. When a negative agitation electrical stimulus is used, a positive pulse may be used to reverse any oxidation (e.g., oxidation of the electrodes).
[0077] Because individual pore protein mixes can have smaller or larger percentages of well-formed, active pore molecules, the desired concentration to use for different batches of pore protein varies. In some embodiments, a particular pore protein mix is first tried on a chip to see how active it is and then the concentration is adjusted until application of the mix will put only a few (e.g., 0-10) pores into an array of 264 bilayer covered electrodes without any stimulation. In general, a concentration may be chosen that results in less than about 10 percent pore formation un-stimulated. In some embodiments, a concentration may be chosen that results in less than about 30 percent pore formation un-stimulated. At such a concentration level, an insignificant number of pores will be inserted when the mix is simply left on the array with no stimulation. At this low concentration, electroporation techniques will insert pores between −100 mV and −600 mV. Positive pulses can be used as well.
[0078] In some embodiments, applying concentrations of alpha-hemolysin or MspA pores in the range of 0.1 ng/mL (nanogram) to 2 μg/mL (microgram) of pore protein will typically result in the preferred condition of having only a few pores inserted in a field of 264 bilayer covered electrodes unaided. The exact concentration used may be determined after calibration on a chip and prior to distribution of the protein pore mix to researchers. In such a case, the resulting protein mix is diluted to the optimal level and then stored for future use and distribution to researchers where further calibration is not necessary.
[0079] The amount or concentration needed to reach the desired state described above varies with salt concentration, temperature, pressure, and the dimensions of the bilayers covering the electrodes. Higher temperatures or pressures require lower pore concentration. Larger bilayer diameters also require a lower pore concentration to successfully implement an electroporation scheme for directed, controlled pore insertion. The pore concentrations above take into account possible variations for salt concentrations (KCl or NaCl) (e.g., from 50 mM to 1 M), temperatures (e.g., from 0 degree Celsius to 25 degree Celsius), pressures (e.g., normal barometric pressure at sea level), and bilayer diameters (e.g., from approximately 5 μm to 250 μm). The technique described above of testing a given mix or representative mix allows for these variances to be taken into account.
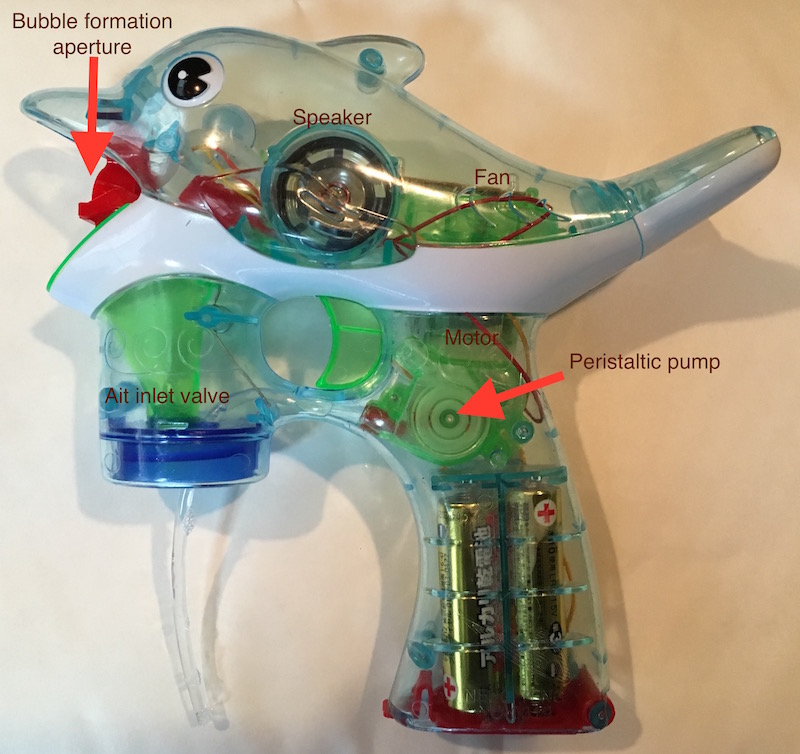
 In order to create bubbles the toy needs not only to deliver the fluid in a controlled manner and allow it to form a film, but to force air through the film in order to create bubbles. This uses the simple (but I still think rather nicely designed) fan assembly shown to the right.
In order to create bubbles the toy needs not only to deliver the fluid in a controlled manner and allow it to form a film, but to force air through the film in order to create bubbles. This uses the simple (but I still think rather nicely designed) fan assembly shown to the right. In order to keeps costs low the toy uses a couple of other neat tricks. The first is the usual ultra-cheap PCBs that are used in these kinds of toys. Single layer and massless. Rather than being CNC routed out I’ve heard these are punched out, given than normal PCBs cost almost nothing, these must be practically free. Again, as is common it uses a dye on PCB with a blob of epoxy on it. This actually isn’t required for the general operation of the toy, it’s only used to synthesise the annoying music the toy produces (and flash the LEDs).
In order to keeps costs low the toy uses a couple of other neat tricks. The first is the usual ultra-cheap PCBs that are used in these kinds of toys. Single layer and massless. Rather than being CNC routed out I’ve heard these are punched out, given than normal PCBs cost almost nothing, these must be practically free. Again, as is common it uses a dye on PCB with a blob of epoxy on it. This actually isn’t required for the general operation of the toy, it’s only used to synthesise the annoying music the toy produces (and flash the LEDs).
 I’m impressed by how robust the toy is. Even though it’s in no way water tight it’s still operational after being fully submerged. The rear LED failed but everything else works. I put this down to the simplicity of the electronics and the fact that the IC is enclosed in resin. But it looks the motor housing maybe more or less water tight. Pretty impressive in any case.
I’m impressed by how robust the toy is. Even though it’s in no way water tight it’s still operational after being fully submerged. The rear LED failed but everything else works. I put this down to the simplicity of the electronics and the fact that the IC is enclosed in resin. But it looks the motor housing maybe more or less water tight. Pretty impressive in any case.