Roswell Biotechnologies
Continuing on with my look at various sequencing companies. Today I decided to take a look at relative new-comer Roswell Biotechnologies.
Business
Roswell Biotechnologies was founded in July 2014. As of Jan 2018 they had about 15 employees and had raised about $6 million in funding from private investors according to Genomeweb [1]. I couldn’t find any SEC filings for the company (if someone can explain this, I’d be curious to better understand when SEC filings happen/don’t happen). The company is based in San Diego.
Technology
The Roswell approach is kind of interesting, it appears to be a tunnelling current detection approach. Other groups are investigating the use of nano gap systems employing direct detection or funcionalised tips (in the interests of full disclosure, I was previously CTO of a tunnelling current detection sequencing company). The Roswell approach however, is quite different.
Roswell still use a pair of electrodes to detect bases, but they build a somewhat more complex system to slowly feed a strand past the electrodes:
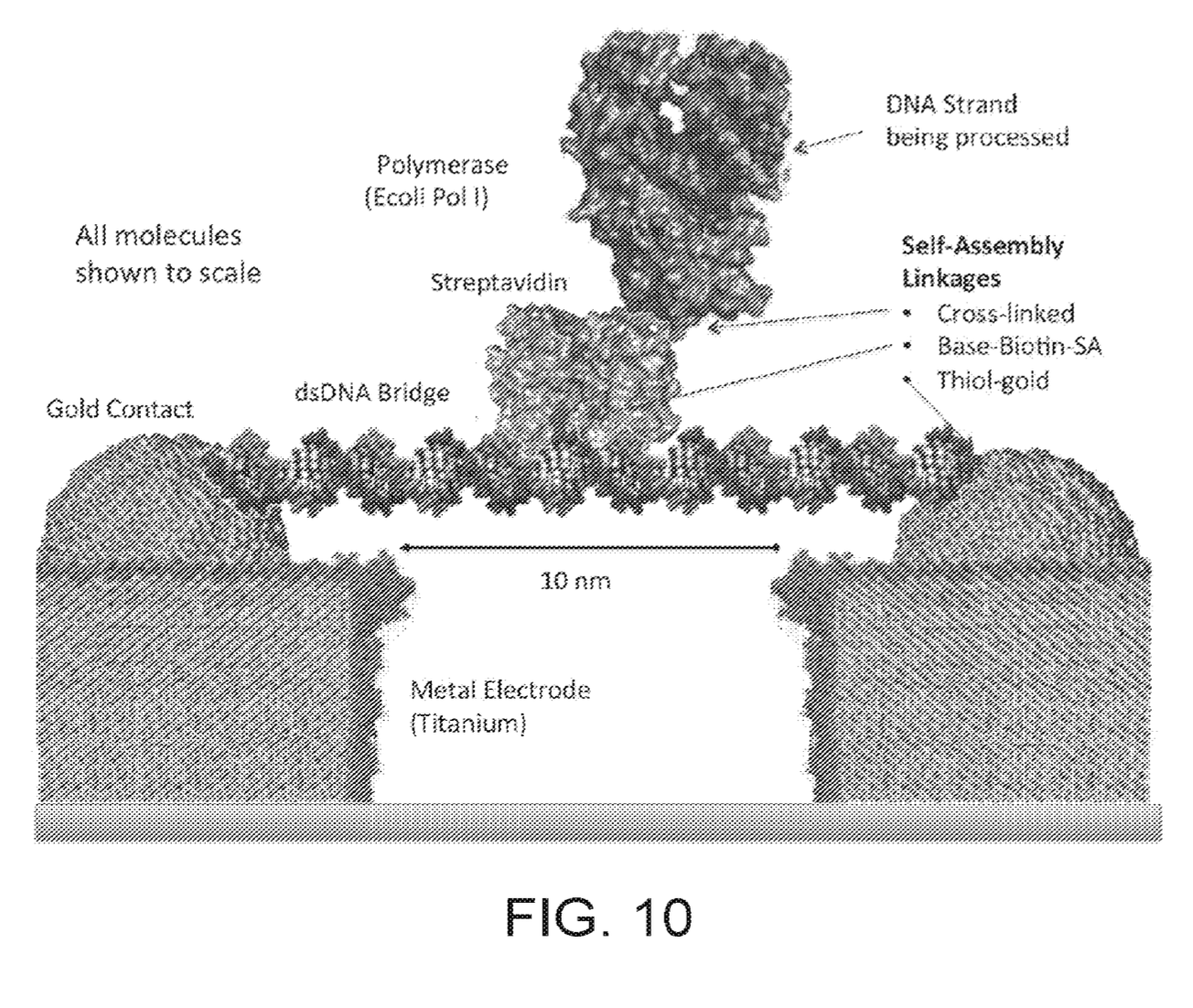
In the above schematic, a double stranded synthetic fragment of DNA is suspended between two gold contacts. This strand isn’t being read, it’s just a scaffold on to which a polymerase is mounted. A second, template strand second comes in (your sample) and is processed by the polymerase.
As bases are incorporated, they interfere with the tunnelling current between the electrodes. I guess the resulting current changes are possibly a combination of interference from the incorporation process itself and the double/single stranded DNA going past the electrode as it is processed by the polymerase. All that really matters is that each base gives a characteristic signal.
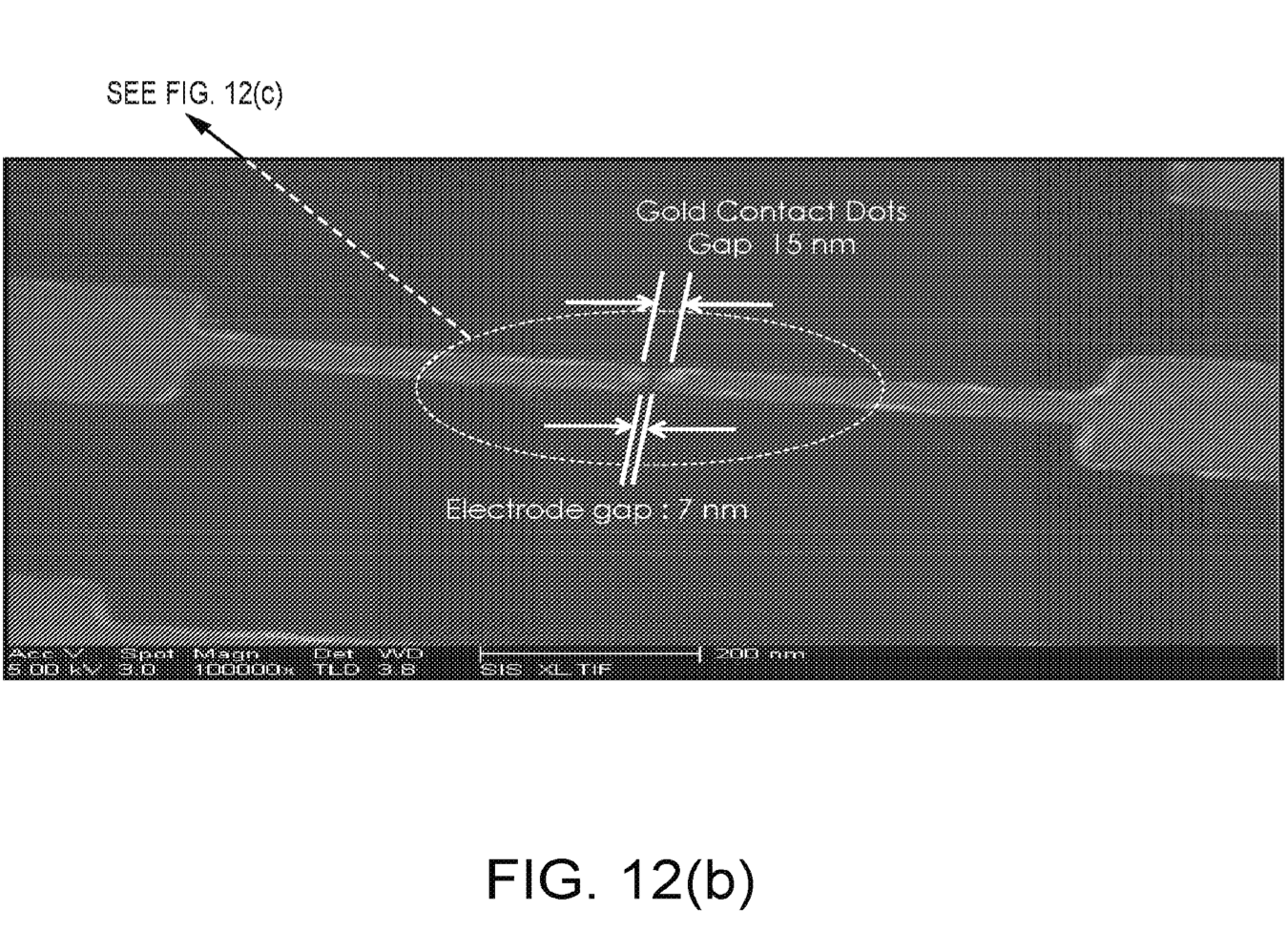
The patent [2] I’ve looked at shows real devices, and includes fabrication details. There are also additional patents which discuss methods of fabricating 18nm gold beads [3] these appear to be used to create gold contacts on which the synthetic DNA support is bound.
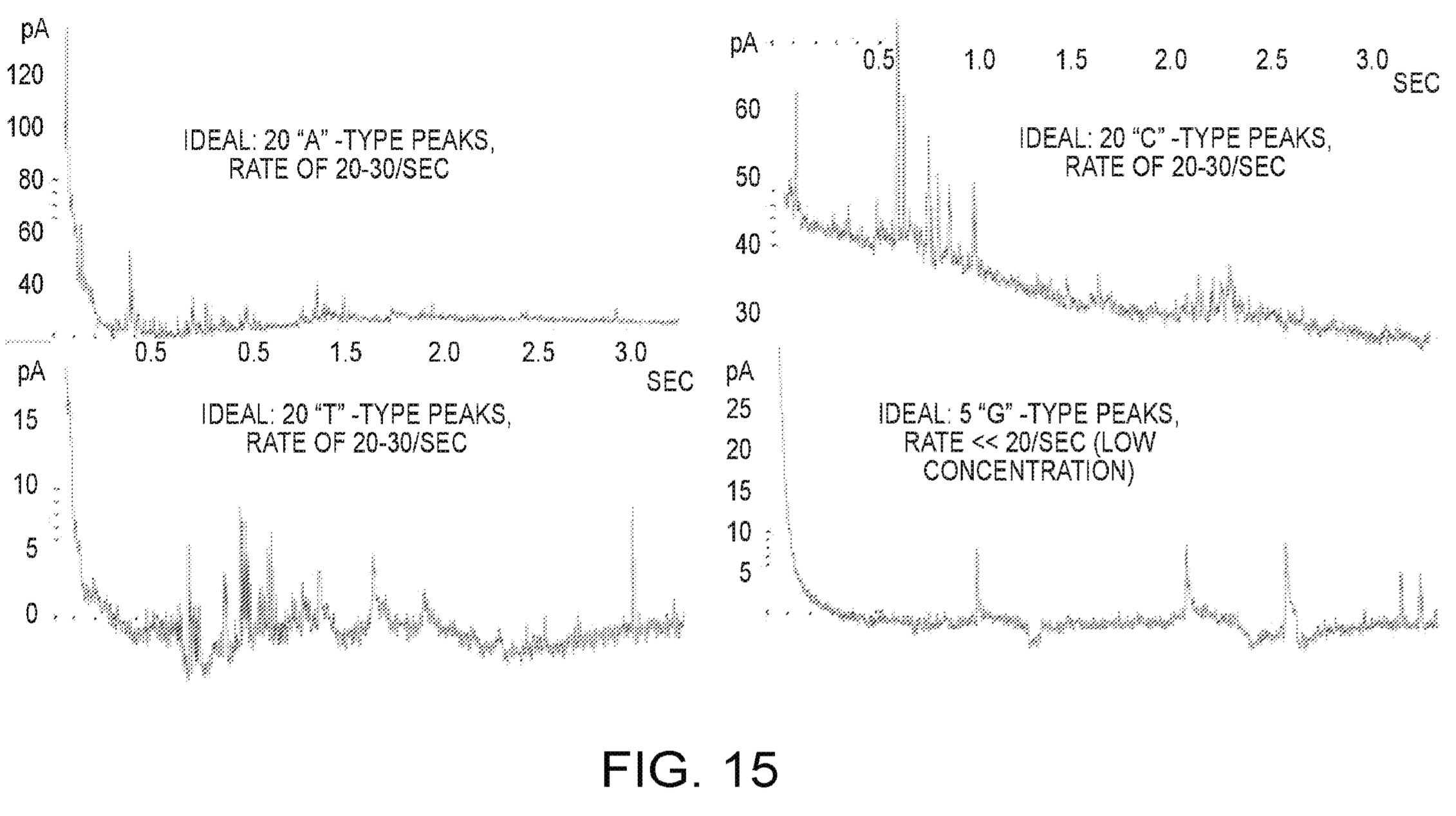
In addition to devices, they also show some data, this appears to be from long homopolymer runs (20 bases). Defections appear to be on the order of 60pA, they say that bases are processed at the rate of ~10 per second.
The graphs are slightly unclear to me, but as I understand it each peak is a single base. So between bases the current returns to baseline.
The approach seems very interesting, and I look forward to seeing more raw data. The genomeweb article says that they’re planning to enter early access testing by Q4 this year [1], and commercialise the system in 2019. So I’ll be keeping an eye out!
Notes
[1] https://www.genomeweb.com/sequencing/roswell-biotechnologies-harnesses-molecular-electronics-chip-based-dna-sequencing
“In Roswell’s approach, a single DNA polymerase is tethered to a molecule, for example a carbon nanotube or a piece of DNA, that is part of an electronic circuit. When the polymerase binds a DNA template and starts synthesizing DNA, the current flow through the circuit changes with each nucleotide incorporated, creating a signal that is base-specific and can also detect base modifications.
“We really have the polymerase wired into the circuit, and you are directly electrically monitoring what it does,” Merriman said. “This actually gives you a much more precise signal of what base is being incorporated than you get from nanopore-based approaches to interrogating DNA.”
The current levels are similar to those used in nanopore sequencing, on the order of picoamps, and the company has so far found no negative effect of the current on polymerase activity.”
“The plan is to have a system ready for testing by early-access users in the fourth quarter and to start commercializing the system in 2019.”
[2] http://www.freepatentsonline.com/WO2017132567A1.pdf
“sequencing genome compatible electrodes with a 5-20nm nano-gap there between, each pair of gapped electrodes having a pair of Au islands, which are for attaching immobilizing biomolecules, such as proteins or fragmented DNA, as bridges across the electrode pairs”
“Such electrode pairs are disclosed herein as usable within a device for genomie/DNA sequencing through electronic conductance measurements.”
“In this illustrated example the bridge biomolecule comprises a synthetic double stranded DNA molecule 20nm in length (60 bases) with thiol groups at both 5′ ends of the molecule for coupling to the Au contact points provided on the metal electrodes.” – Figure 10
With reference now to FIG. 15, the measuring of incorporation signals using the sensor is demonstrated. The plots in FIG. 15 show the current signals resulting from the sensor being supplied with various primed, single stranded DNA sequencing templates and dNTPs for incorporation and polymerization. In each case, the major signal spikes represent signals resulting from discrete incorporation events, wherein the polymerase enzyme adds another base to the extending strand. In the plot at the upper left, the template is 20 T bases. In the plot at the upper right, the template is 20 G bases. In the plot at the lower left, the template is 20 A bases. Lastly, in the plot at the lower right, the template is 20 C bases. The approximate rate of incorporation observed is about 10 to 20 bases per second, which is consistent with standard enzyme kinetics except for the lower rate of ~1 base per second presumed due to rate limiting factors (e.g., lower dNTP concentration).
[3] Fabrication of 18nm gold beads: https://patentimages.storage.googleapis.com/64/98/90/9ed2d36b40e4dc/US20170240962A1.pdf