Depixus (was Picoseq)
Business
Depixus (originally Picoseq) was founded in 2012 in Paris, based on IP developed by Vincent Croquette, David Bensimon, and Jean-François Allemand at École Normale Supérieure (ENS). In 2013 the company was seeking 5M Euros, but it’s not clear to me if the round closed [1]. In May 2016 Arix Biosciences agreed to acquire up to ~20% of the company for 1.24M Euro (paid 0.93M Euro, with it appears the rest relying on milestone payments, though reports vary) [2].
While the company is based in Paris, a Cambridge (UK) branch exists. Full accounts appear to be filed in the UK [3] and it seems that at the end of 2016 they had around 1M Euro is cash, and 800K Euro in cash to come. In 2016 operating costs were listed at 1.3M Euro, personnel costs at ~600K Euro.
Technology
The technology behind Depixus is described in a 2012 paper. Fundamentally the detection process relies on the preparation of hairpin’d double stranded DNA (double stranded DNA with a loop on the end). One end of this strand is connected to a surface, the other to a magnetic bead.
The bead is held in a magnetic field that can be varied. By varying the field the bead can be moved, pulling the DNA. You can therefore pull the double stranded DNA apart “unzipping” it. It’s also possible to reduce the field strength, and allow the DNA to zip back up. While applying this force, you can also sense the location of the bead. In the 2012 paper this is performed optically, by analysing the diffraction rings around the illuminated beads:
This system allows Depixus to measure the forces required to zip/unzip the DNA, and it is this measurement, and interference with the zipping process, that Depixus uses to sequence DNA.
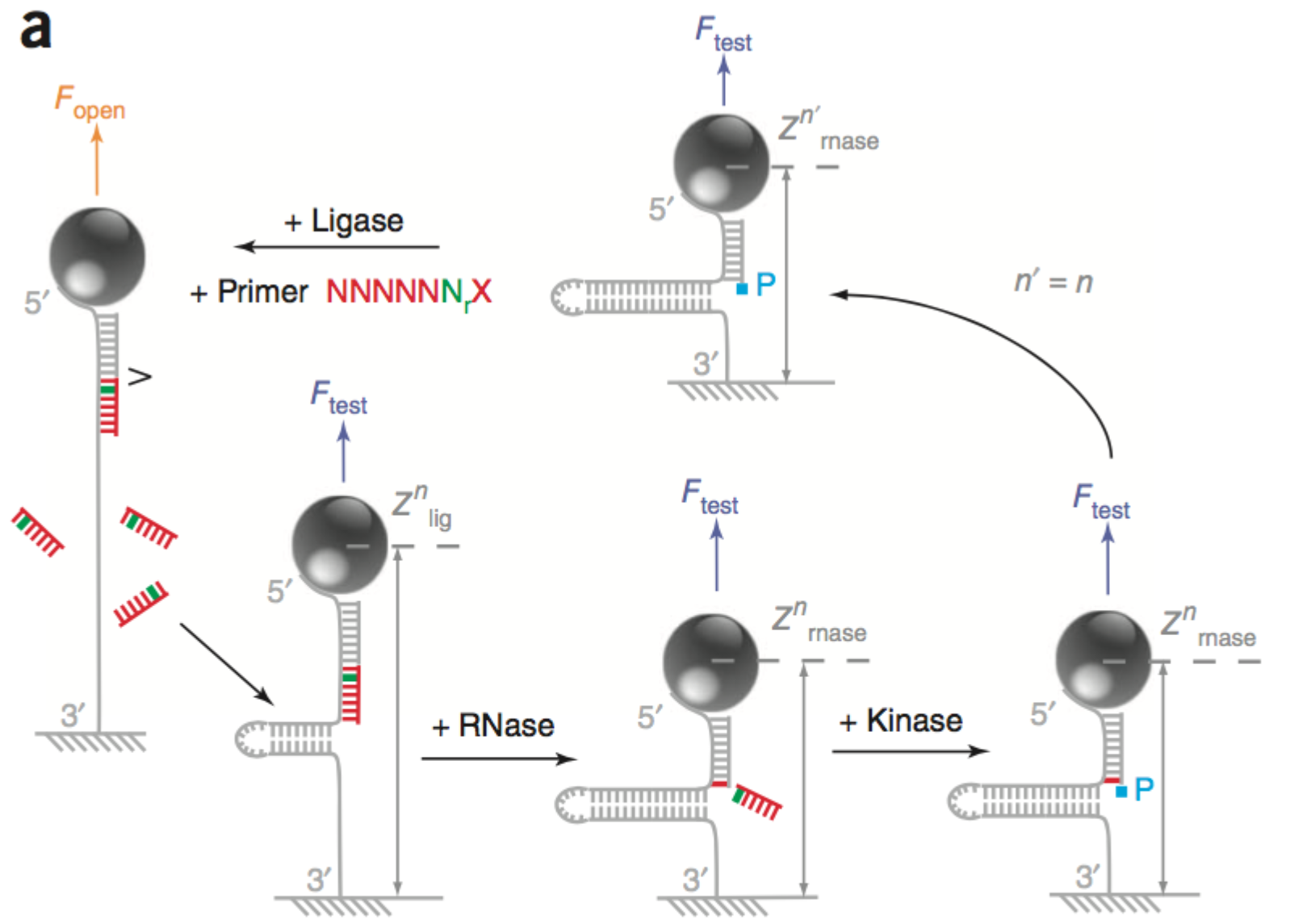
In the 2012 paper [5] they show a hybridisation and a ligation scheme (illustrated in the schematic above). In this scheme they fully unzip the strand then ligate a probe (7mers, all Ns expect for a single known location). They then allow the strand to zip up again. If ligation was successful, the bead position is offset by ~5nm. This change in position, tells you if the probe ligated, and therefore which base is present at the known location. They then cleave off 6 nucleotides (if ligation was succesful) and perform the process cyclicly to determine the complete sequence of the strand.
I briefly looked for additional progress on the platform since the 2012 paper. The website currently suggests that they are able to detection base modifications, but I was unable to determine the exact mechanism. One report on their Arix deal, suggests they have generated 286bp reads [2], but again provides few details.
A patent from 2016 [4] covers an improved detection system. Rather than imaging the beads directly they sit the beads in a well. The solution is conductive (contains ions I guess), and there is a bias voltage from the bottom of the well, into the solution. As the bead moves up and down it blocks the aperture of the well. This blocks the ionic current flow, effectively varying the resistance. So, you can measure that current blockage at determine how much the bead is blocking the aperture, and therefore its location.
I’ve not yet been able to find other patents covering the detection of modified bases, it’s possible that they’re not yet available or that they can be found with further investigation.
The technology is quite interesting, in some ways it reminds me of Cearus (detection via changes in beads location/movement) and Complete (ligation). As always it will be interesting to see how things progress.
Notes
[1] Genomeweb article. https://www.genomeweb.com/sequencing/picoseq-expanding-potential-applications-hairpin-dna-based-technology-it-enters
“The company is currently seeking funds from a variety of sources and/or potential partners, PicoSeq CEO Gordon Hamilton said, and hopes to reach its current target of roughly €5 million ($6.6 million) late this spring or sometime over the summer.”
…
“Even so, Hamilton emphasized that he does not see PicoSeq as a direct competitor to existing or anticipated next-generation sequencing instruments. Rather, he argued that the mechanical hairpin method might have a range of applications that are distinct from — or complementary to — those offered by such platforms.
For instance, because each DNA hairpin can be opened and closed tens of thousands of times, it should be possible to look at the same molecule multiple times — perhaps searching for a DNA fingerprint or barcode in one instance and profiling a full sequence or a suite of specific epigenetic modifications in another.
“You could potentially build up the level of detail that you want from the picture based on your requirements as a user,” Hamilton said, “rather than taking the kind of sequencing approach where you chuck everything in and sequence the whole lot and deal with the data afterwards.”
[2] Various references to Arix Biosciences investment.
https://propertibazar.com/article/prospectus-arix-bioscience_5a4f5e8fd64ab2089e446872.html
“On 6 May 2016, Arix Holdings entered into an agreement with Depixus pursuant to which it acquired an interest of approximately 21 per cent. of Depixus’ share capital for an aggregate amount of €1.24 million, paid in three tranches upon achievement of certain technological development milestones in May 2016, December 2016 and July 2017.”
https://www.researchpool.com/download/?report_id=1284000&show_pdf_data=true
“Depixus is a laboratory technology company that is developing novel methods of sequencing DNA and RNA that reveal epigenetic information in addition to the nucleotide sequence. In May 2016, Arix entered into an agreement to acquire up to 18% of the company for €1.24m, of which €0.93m has been paid (for an assumed current stake of 13.5%).”…”The technique is performed by attaching a small magnetic bead to a hairpin of DNA immobilised on a solid substrate and then using magnetic force to unfold the hairpin. A soluble oligonucleotide is added to the solution and the strand is allowed to refold. If the oligonucleotide binds to the DNA strand, it stalls the folding process and alters the force between the bead and magnet. Using a series of oligonucleotides in this way, a complete sequence can be determined. The longest complete sequencing read that has been reported was 286 base pairs, but much longer sequences (over 20,000 base pairs) have been interrogated using the technique, posing the possibility of significant read lengths.”
Arix Bioscience invested in Depixus:
https: //www.researchpool.com/download/?report_id=1284000&show_pdf_data=true
https://www.google.com/url?sa=t&rct=j&q=&esrc=s&source=web&cd=26&ved=2ahUKEwi-1Irst8TcAhVLurwKHXh2CWE4FBAWMAV6BAgFEAI&url=https%3A%2F%2Fwww.gov.uk%2Fgovernment%2Fuploads%2Fsystem%2Fuploads%2Fattachment_data%2Ffile%2F725229%2FInnovate_UK_funded_projects_from_2004_to_1_July_2018.xlsx&usg=AOvVaw3FUMKtIp67PHBWQojQTJ4E
[3] https://beta.companieshouse.gov.uk/company/FC032917/filing-history
Accounts as of end of 2016, appear to have had 1M Euro in cash in hand, and 800K Euro to come.
[4] Patent from 2016. It’s assigned to the research institution and Depixus CEO rather than Depixus directly, which seemed a bit weird to me. But it appears to be associated with the Depixus technology. https://patents.google.com/patent/WO2016177869A1/en?oq=Gordon+Hamilton+sequencing
[5] 2012 paper: https://www.nature.com/articles/nmeth.1925