Ultima Genomics
Not much is publicly available regarding Ultima genomics. The company appears to have received >3M USD in SBIR awards in 2019 and 2020 (which is by usual standards, rather a lot). Its CEO is Gilad Almogy who was previously CEO and founder of Cogenra (a solar company). Aside from this I can’t find much information about fund raising. The company appears to have been founded in late 2016. From the SBIR application, and patents it seems clear that they are focused on DNA sequencing.
Technology
I’ve taken a very brief look at a couple of Ultima’s patents. The first patent describes an imaging system for use in with a DNA sequencing platform. The substrate is on a rotating platform:
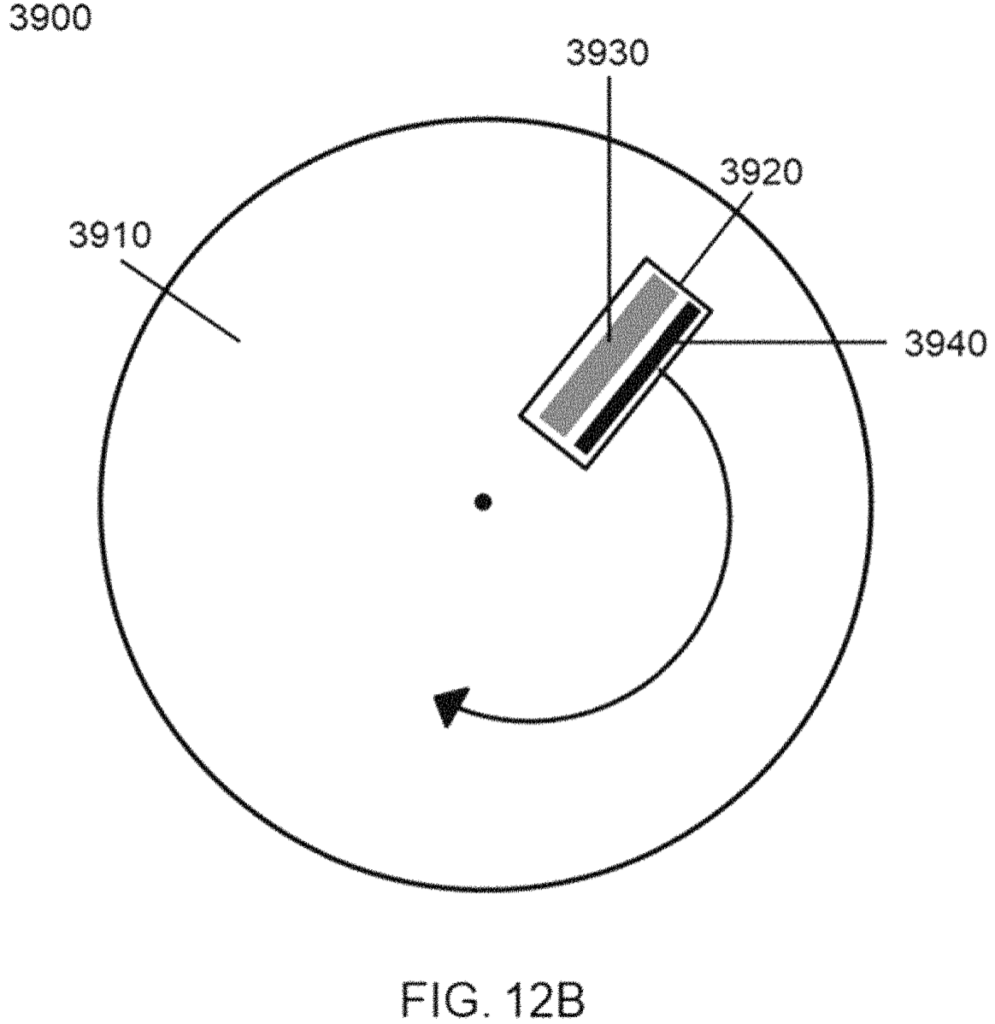
Which is somewhat reminiscent of Ling Vitae approach. Where a fluidic system was incorporated as part of a CD-like platform. I kind of like this idea, because you can potentially image a large area, with one stable axis. After an imaging pass, you end up where you started, ready to image again. In a cyclic sequencing approach, this seems attractive.
The patent also shows what looks like real image data from an ordered array:
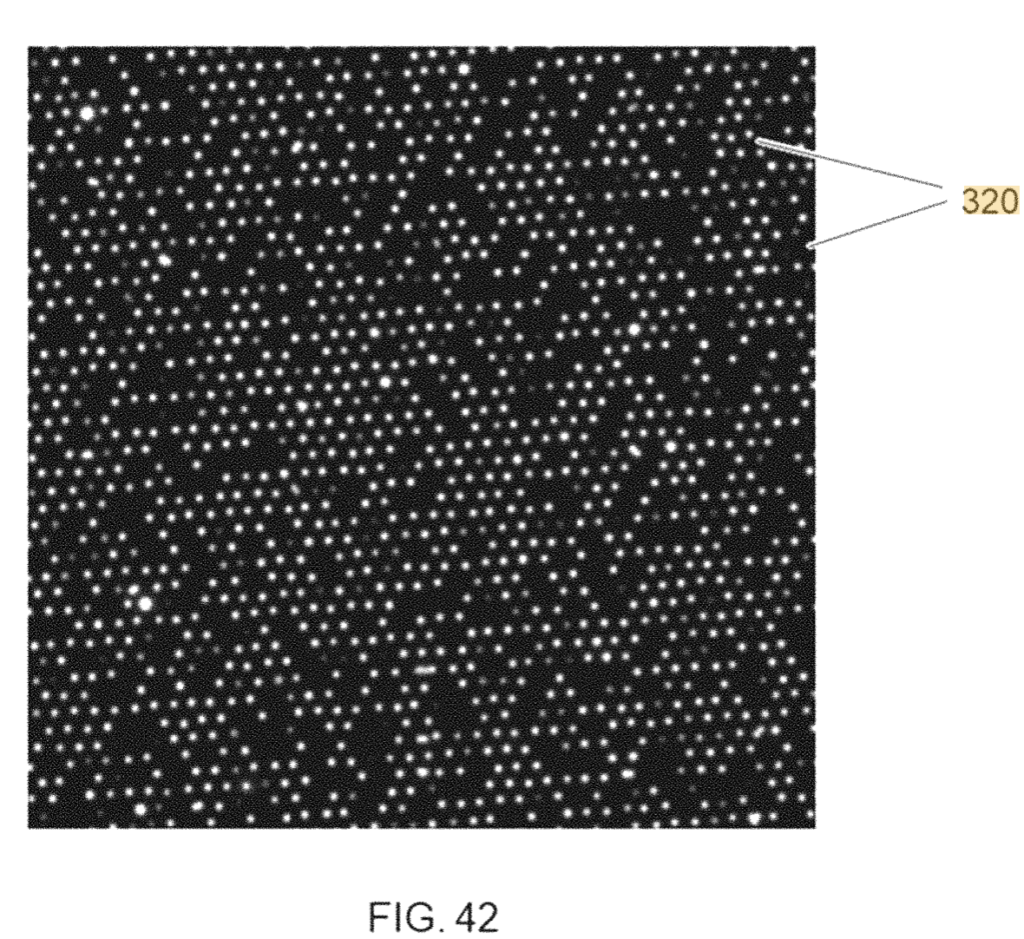
And even some read statistics. I’d need to read this in more depth, but I suspect this is just extrapolated from spot counts:
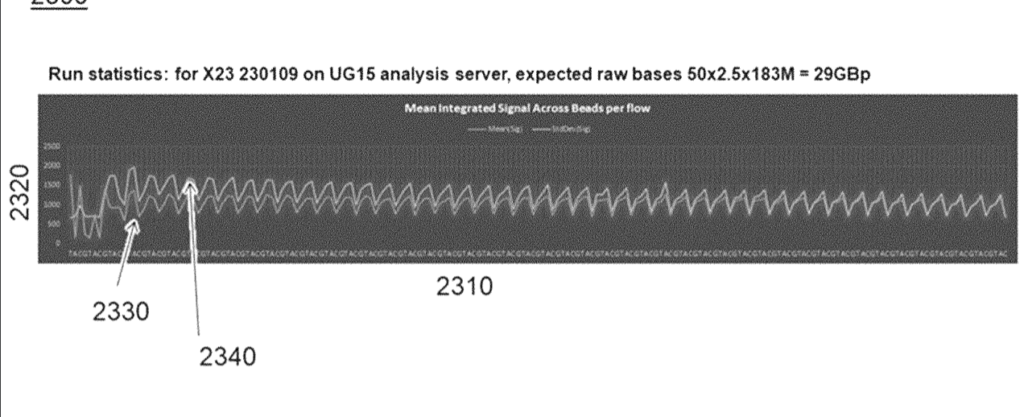
The second patent describes a sequence-by-synthesis (SBS) sequencing chemistry. The approach appears to be somewhat similar to standard SBS, in which nucleotides are flowed in and detected in cycles. However here (unlike Illumina) reversible terminators do not appear to be used.
This is also not a traditional single-channel/unterminated SBS platform (Ion Torrent, 454), which would incorporate a single base type in each flow.
The Ultima Genomics approach presented here, works on a population of templates (so a surface amplification approach like cluster generation, or a bead based approach would need to be used).
In the dominant embodiment, it appears that Ultima Genomics would incorporate mixtures of terminated and unterminated bases. Most positions would extend normally using a wildtype nucleotide. However a smaller fraction of nucleotides would be terminated, preventing templates from extending any further. These terminated nucleotides would be labelled (likely a fluorescent label). Allowing the sequence of a cluster to be determined from the terminating sub-population.
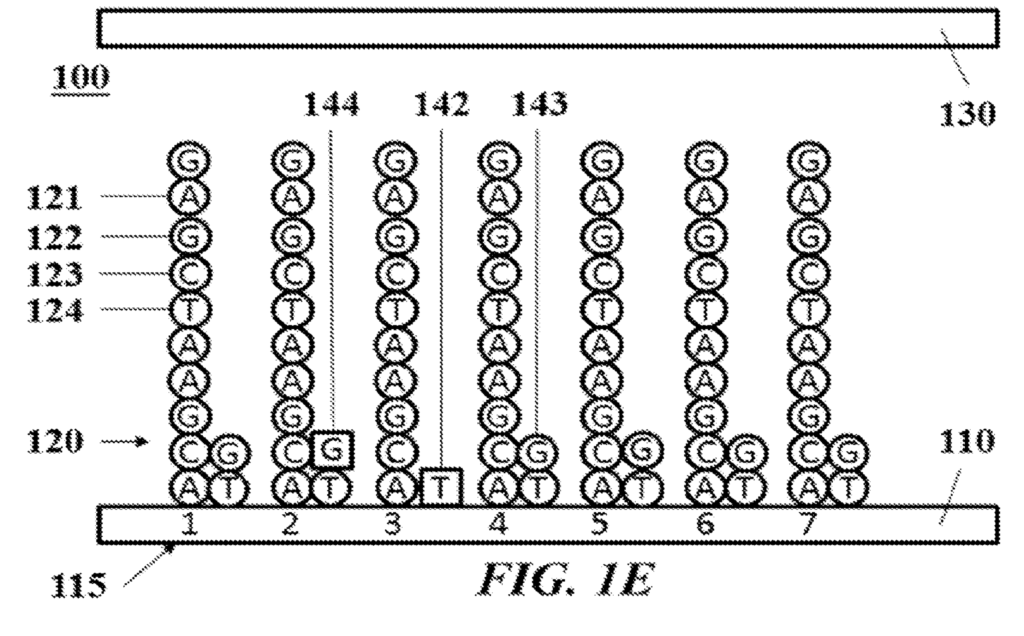
One obvious issue with this approach, is that with each cycle, you get an accumulation of incorporated dye. You could cleave the fluorescent labels, which seems like the obvious thing to do. But the patent suggests a different approach, essentially just monitoring the increase in fluorescent intensity. An increase in a cycle means that a base was incorporated, no increase – no incorporation.
That’s fine as it goes, but eventually the build up of dye is going to make imaging problematic. You’ll either overflow the cameras range, or get increased crosstalk from scattering etc.
To avoid that one suggestion in the patent is to periodically switch dyes. That is just move to a different dye with a different emission after a few cycles. You can then switch filters, effectively cutting out fluorescence from the accumulated dye.
Overall some aspects of this approach don’t seem particularly new. Using terminators was of course the foundation of the original Sanger sequencing approach. And mixtures of nucleotide types have previously been used for sequencing in academic work. Some of this prior work may explain why the majority of claims on this patent have been cancelled.
The chemistry itself seems more complex than those currently available… so what is the advantage? I suspect the motivation is that wildtype bases likely incorporate more efficiently. This might give some improvement over Illumina-style SBS. The fact that platforms using wildtype nucleotides (Ion Torrent, 454, Sanger) have somewhat longer read lengths (on the order of 1000 nucleotides, versus 150) supports this.
But in this Ultima approach, the loss of templates through termination will eventually limit read length. It’s also worth noting that while the other platforms mentioned above have longer reads (and in some cases lower error rates) they have not proved commercially very successful. So I’d be concerned that the added complexity of the Ultima approach doesn’t provide enough benefit over existing platforms.
That said, this is only a quick review of a couple of patents. I will be watching with interest to see how things develop.