SomaLogic
This post previously appeared on the substack.
Somalogic are developing a aptamer based protein detection platform with a microarray-like readout. Aptamers have been around since the 1990s, the basic methods being far older still. The basic idea is that you evolve a DNA or RNA sequence that will specifically bind to a protein of interest. Aptamer’s have a troubled history and I’m not really aware of many successful commercial applications based on aptamer approaches.
Somalogic claim to have solved the aptamer problem by adding 4 additional modified nucleotides to the existing natural set. The basic method was published in 2010. This gives them 8 basic monomers to work with (still a lot less than 20+ amino acids, but in the ballpark of the ~10 that have been proposed as a minimally viable set).
The heart of this paper shows that they can find aptamers for various proteins using modified nucleotides where they previously couldn’t:
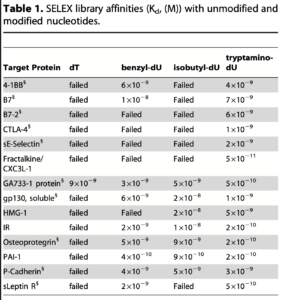
Kd values here range from 10⁻⁸ to ~10⁻¹⁰ (smaller is better). This in the range of high affinity antibodies (but not very high). These values also compare well to those for regular aptamers, for example these in with 10⁻⁸ to 10⁻⁹ range.
However even antibodies have a relatively high false positive rate (for example, 0.4% for COVID19 tests). A false positive rate of 1 in 250 may not seem like much. But when you’re looking at ~10000 proteins, you’ll get 40 false positives. Of course, this doesn’t exactly map to the Somalogic approach where you’re looking to quantify expression levels rather than obtaining a yes/no answer.
So, particularly coming from a background in genomics, the Somalogic readout is likely to be noisy. This is likely the case for any large-scale aptamer/antibody platform. Olink, by linking two affinity reagents may be able to reduce this. Nautilus, by using multiple affinity reagents sequentially, may also have a route to a less noisy readout.
But what Somalogic may have, is more resolution on their readout. Unlike Olink and Nautilus, the Somalogic platform doesn’t appear to be using a single molecule approach.
Essentially they fish out the aptamers that bound, and then pull these down onto a microarray like platform (hybridizing the aptamers to probes on the surface). Microarray’s (and hybridization) always seems like a process fraught with issues, with environmental issues (like temperature and ozone) effecting results. All issues, you rarely see with sequencing.
So, there’s potential for binding issues between aptamer-protein and with the aptamer-array. The upside is that the array gives you a large dynamic range (I believe they suggest 12 orders of magnitude). So rather than having to readout billions of molecules (as Nautilus or Olink might) you can get this information from the Somalogic platform by imaging each protein (aptamer type) once. I.e. an array with on the order of 10,000 spots. A fairly small (and likely cheap) array by DNA microarray standards.
Final Thoughts
Somalogics secret sauce seems to be in their modified nucleotide aptamers. I would guess at best these are one order of magnitude better than regular aptamers. So you can make the argument (like Nautilus) that you could just use a couple of lower affinity aptamers rather than one slightly higher affinity one.
Aptamers do give Somalogic a natural way to generate a high dynamic range readout (using microarrays). That’s neat, but brings its own issues. And overall, I suspect compensating for artifacts may be harder than in single molecule platforms (like Nautilus and Olink).
All these platforms are likely to have issues with binding affinity, background signal, and other environmental factors.
These are issues that a true protein sequencing platform could potentially overcome.