Notes on Genia’s new paper – nanopore SBS
 Genia have released a new paper showing recent data from their “nanopore SBS” platform.
Genia have released a new paper showing recent data from their “nanopore SBS” platform.
Summary: The best data in this paper is a 20bp read on a synthetic template with no homopolymers. This has long dwells (multiple seconds) and levels looks clearly differentiated. The second has short dwells (100ms?) under different experimental conditions they say gives better resolution on homopolymers.
The first dataset looks like reasonable progress, the second I’m not sure I buy, and is very low complexity in any case (just 3 homopolymers runs).
Overall this is an R&D level system. It’s interesting progress, but not useful for any application at present.
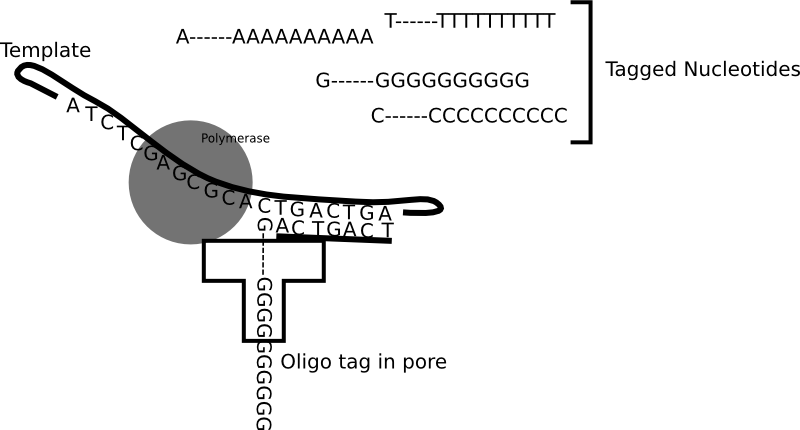
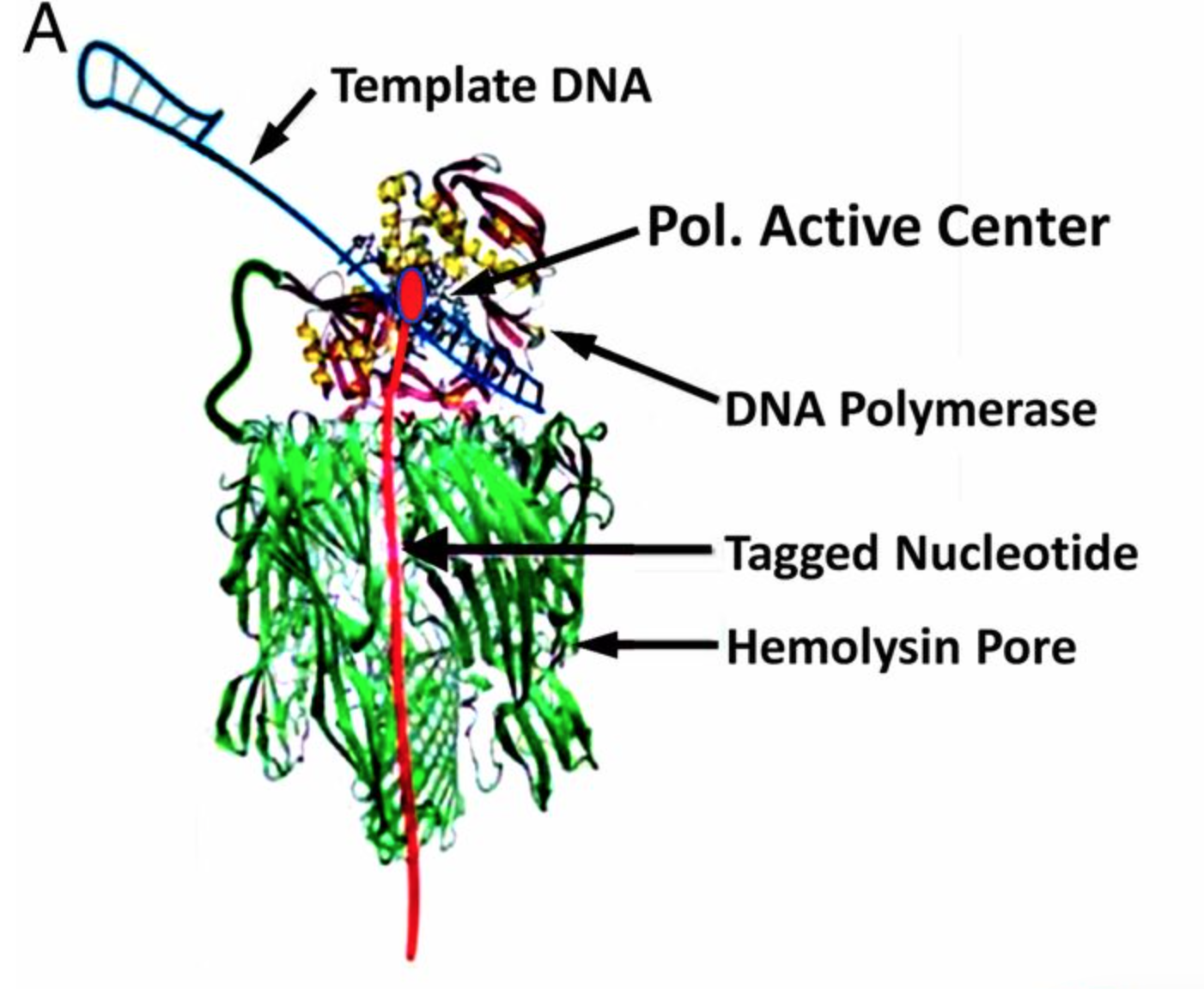
Genia’s nanopore SBS technology is shown in the figure above. To a computational scientist like myself it seems like an interesting system. Genia have had modified nucleotides created such that each nucleotide has an oligo hanging off it. That seems pretty amazing, but it appears that the nucleotides are incorporated by the polymerase. During the nucleotide incorporation process, the tag breaks off and passes through the pore. The cartoon below shows the basic idea:
The diagram above shows each nucleotide tagged with a longer oligo which goes down the pore. When the tags sit in the pore they block the flow of ionic current through the pore. While in the diagram above I show polyN tags, Genia have selected tags to give a good spread of current blockages (and have included modified bases in the tags). Uses oligo tags has two benefits over competing systems. Firstly, each tag is providing a signal from a single base. In some competing nanopore systems multiple template bases are in the pore at the same time. This means that more than one base effects the readout. This results in a convoluted signal from which it can be difficult to extract the original template sequence [1]. The second advantage is that you can optimise the spread of the tags so that each tag and be easily differentiated.
There seems to be one other trick in the system described with this sentence “The applied voltage is adjusted to ensure that, in a majority of cases, one and only one pore is inserted into the membranes of each well. “. My understanding was that the number of pores in each membrane is poisson limited (see new post on this). But if they’re able to control the pore insertion with an applied voltage that’s pretty neat (perhaps someone who understands this can comment). The paper discusses a 264 pore chip, which stuck me as odd as I believe they’re talked about chips with many more pores.
Data
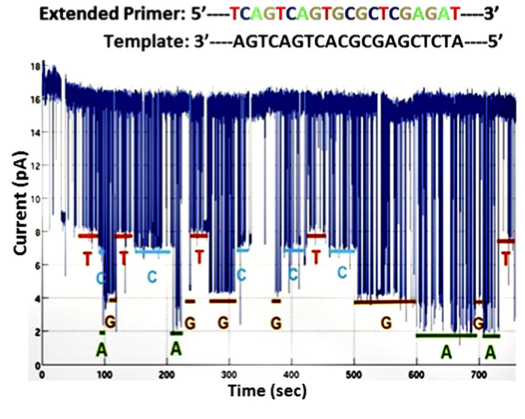
The first dataset is shown to the right. This is the dataset that contains no homopolymers runs. To my mind it’s the most convincing dataset in the paper. Raw data for this plot isn’t available (why is that still ok in 2016?). So we’re forced to draw our conclusions from eyeballing the data, and their analysis.
The data however looks quite clean, I’d assume this is the best data they’ve seen on the chip, and it’s a shame their aren’t more examples of this read. The base dwells seem to be all over the place, and I’d assume, much like other nanopore systems, they are exponentially distributed.
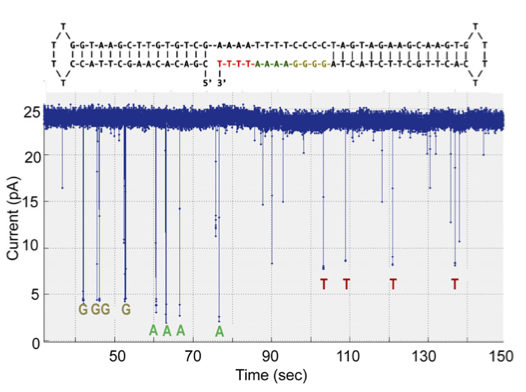
The second dataset describes their experiments optimising the system for homopolymer detection. I find this less convincing. It’s a short run, and it’s hard to tell how much longer the ‘T’ calls are than the noise spikes that appear to be at almost the same level. The following statement also gives me some concern:
“Base calling was carried out by manual inspection of the current level of each deflection, ignoring ones with dwell times less than 10 ms.”
I guess this is effectively thresholding the data, but in that case why not say that? Regardless the fact that an automated base caller wasn’t used most likely means that datasets are very small at the moment.
Overall this is interesting progress and represents a solid milestone in their development. It’s not clear that this actually represents the state of the art Genia system. It may be that this is an older platform and doesn’t reflect the current system, as the low pore count might indicate. However, it’s common for every vendor to say this when a new paper is released, and it’s difficult to discern the truth without further disclosures.
[1] In the pore used here this is particular important. You have about 15 picoamps between the maximal and minimal blockage. This isn’t a huge amount of signal. Before even considering thermal noise, if we were to sample at 1MHz 1 picoamp would be 8 electrons per timestep. As a colleague used to say… so few electrons that you could name them.
Disclosure/Disclaimer: I have worked and continue to work in the DNA sequencing industry. I own stock in DNA sequencing companies. While I have tried to be unbiased this represents my opinion and speculation only. I recommend reading the publicly available paper for yourself.