Nautilus Prospectus Review
This post originally appeared on my substack.
My previous post on Nautilus covered their published IP and was originally released to paid subscribers. After it was released publicly, someone forwarded me the Nautilus’ prospectus. While the information in the prospectus is largely in line with that presented previously, it provides more concrete information on their implementation.
In the prospectus they describe “scaffolds” these are likely the SNAPs (nanoballs) from their patents. The scaffold prep process sounds non-trivial as they state flowcell/sample prep takes ~2 hours.
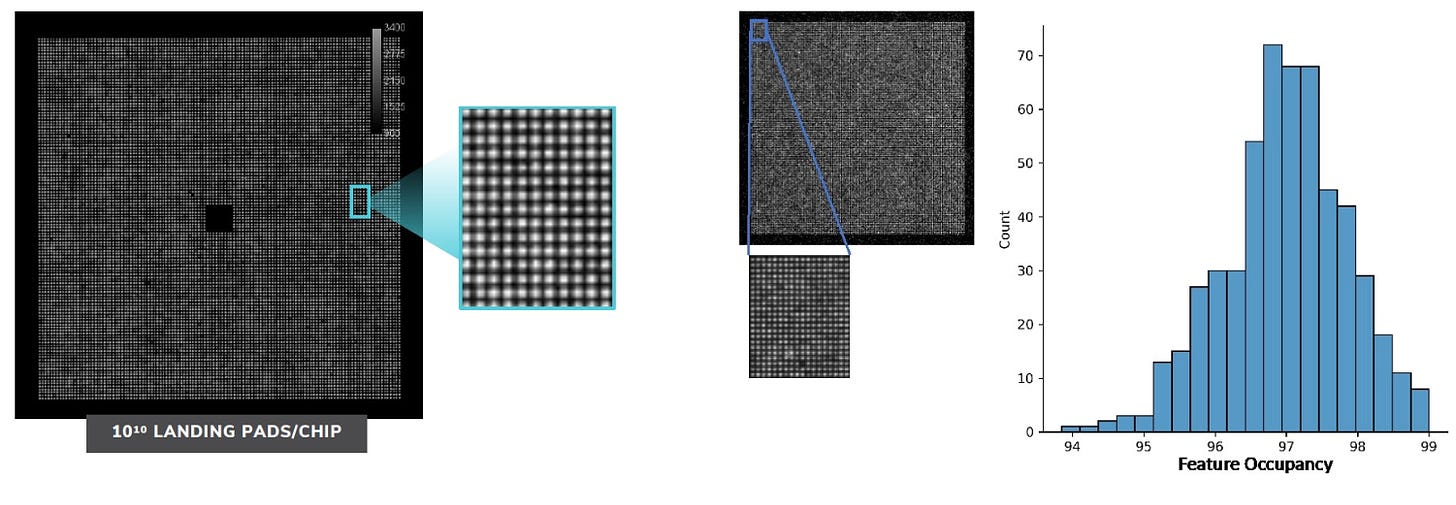
They also make it clear that their current chips have 10 billion sites. This is interesting because it helps frame the value of their arraying approach. To put this in context the Hiseq 2500 was able to reach 4B reads without a patterned flowcells/arraying. So, while the arraying technology is interesting it still doesn’t seem of fundamental importance to getting the platform working. The arraying approach seems to work well though, with >95% of sites being active:
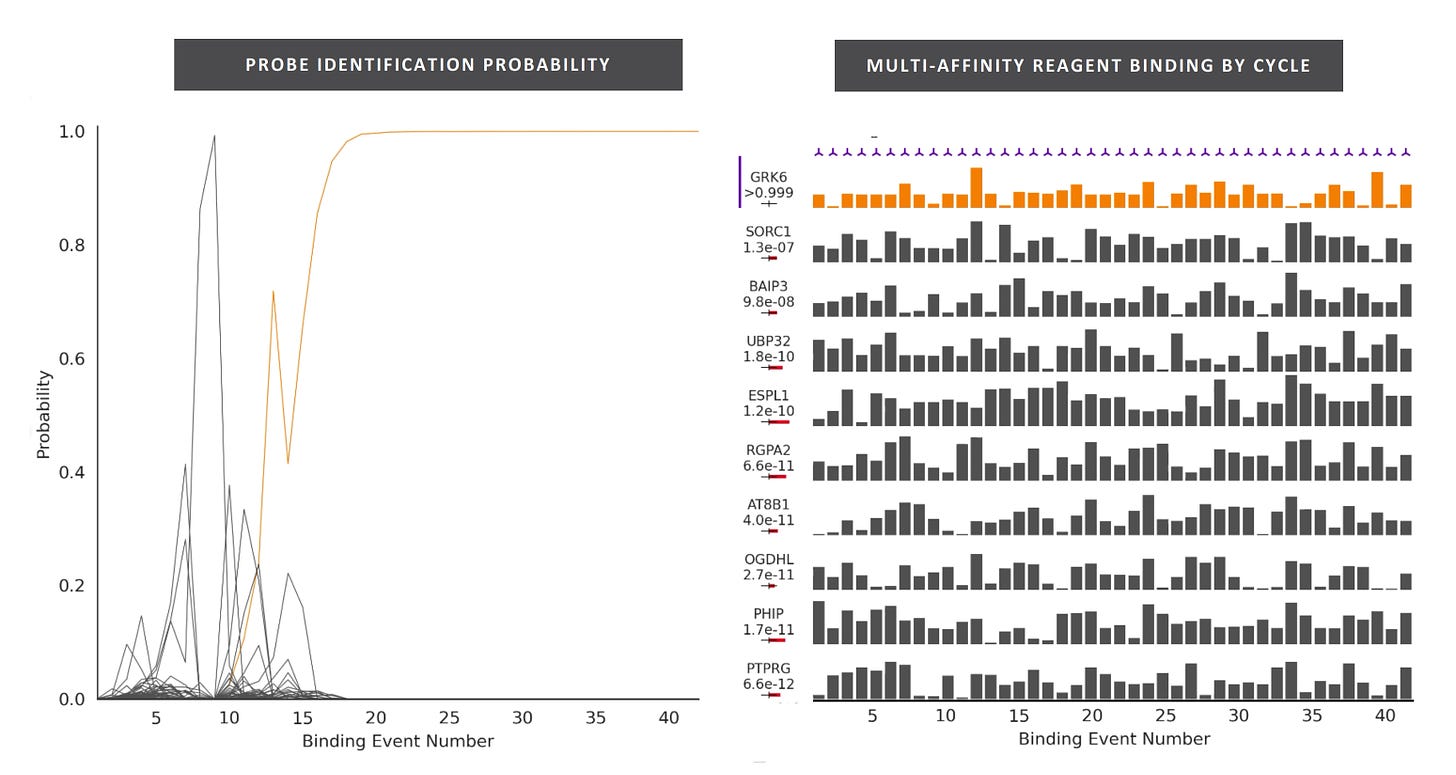
There are a couple of statements on cycle requirements in the document that appear slightly contradictory. The first is “A typical 300 cycle run will generate approximately 20 terabytes of data” the second is “it takes roughly 15 cycles of multi-affinity probe biding events to uniquely identify a protein”. If you only need 15 cycles, why do you typically run to 300?
They present this plot showing classification of a protein using multi-affinity reagents:
My guess is that there are multiple sets of multi-affinity reagents. So it’s more like out of this set of 300 reagents we can find 15 that will give a unique signal for a particular protein. If this is true, then the 15 cycles statement isn’t very meaningful. And it sounds like you need to do 300 cycles in general. This implies a complex reagent cartridge and fluidics system. They state that “Nautilus intends to utilize over 300 complex reagents and various antibodies” which backs this up.
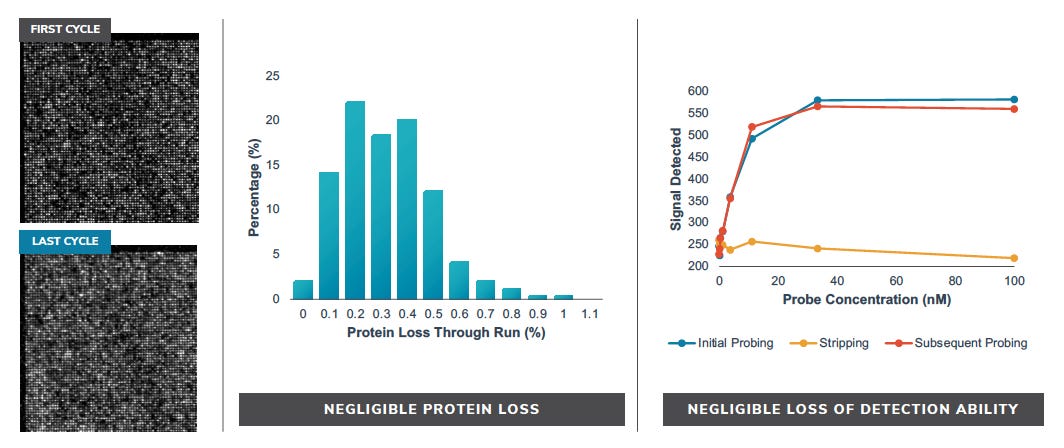
But it seems like proteins stay attached though these long runs (<1% loss):
Beyond this, they don’t say anything regarding affinity reagents other than they can use a “wide variety of “off-the-shelf” affinity reagents” and “we have developed a proprietary process for high throughput generation and characterization of multi-affinity probes”. I’d guess the high throughput generation approach is likely the sequencing based aptamer evolution approach described in their patents.
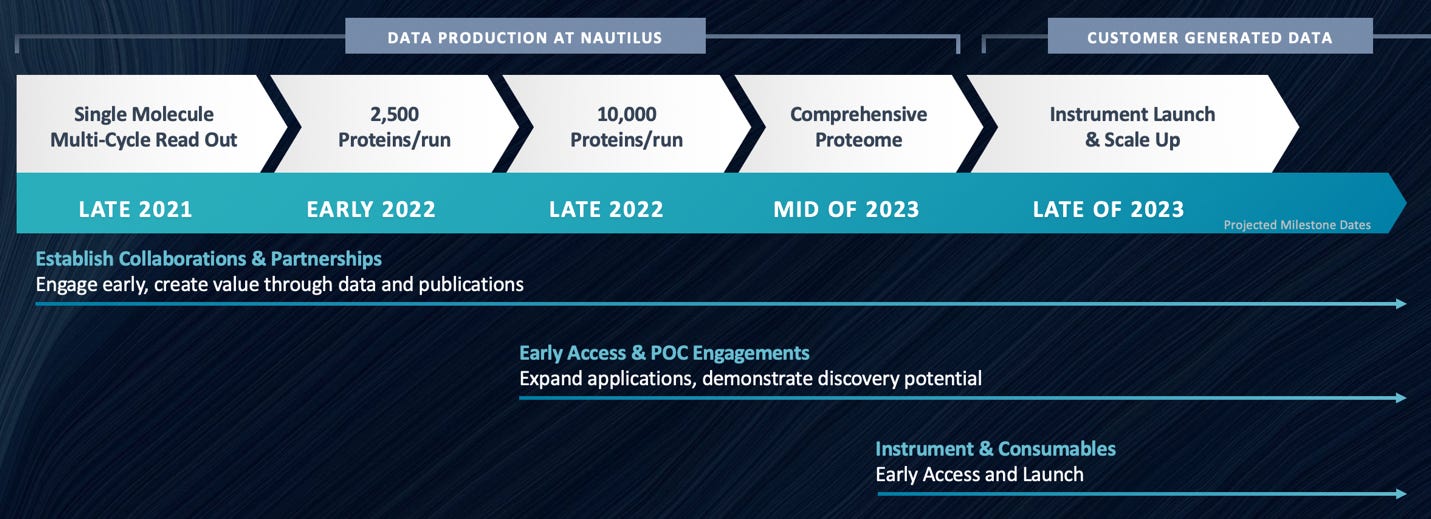
On the commercial side, they appear to be targeting a 2023 launch. So it will likely take some time for us to find out how well the platform works in practice.
Pricing is less clear but they say it will be “in-line with mass spectrometry system budgets allocated for broad scale proteomics applications, and thus with a premium instrumentation average selling price”. However I suspect the consumables pricing will look pretty different to mass spec. That 300 reagent cartridge and patterned flowcell doesn’t sound like it’s going to be cheap.