Base4 Innovation Ltd
Following on from my list of sequencing companies I wanted to delve into Base4 next. Below are my brief notes on their business and technology.
Business
If you’re not really interested in the business stuff, skip this and head to the tech section below. Suffice it to say they’re a series A/B stage DNA sequencing startup and have received about 21M GBP in total investment.
The company was incorporated in October 2007. Being a UK company there’s quite a bit of information at companies house [3].
Investors appear to include: Longwall Ventures, Meridian Corporate Finance, Oxford Technology Enterprise Capital, Royal Society Enterprise Fund, Torteval Investments Ltd, and Amadeus RSEF. Amadeus RSEF I think stands for Royal Society Enterprise Fund, and possibly indicate that this investment is associated with that fund, rather than the main Amadeus fund.
Companies house accounts list 36 staff in 2016 [4] , with a burn rate of about 3M GBP a year. They had around 1.6M GBP in cash. The accounts state “to meet the company’s ongoing cash needs…additional funding will be required…within the next 12 months”. And their site states they received 5M GBP in November 2017 [1]. This may indicate that further fundraising would need to take place in 2018.
Glassdoor reviews [2] for the company contain some entertaining gossip. It looks like there has been some internal restructuring at the company, but with the raise last year hopefully things have settled down.
Technology
The image below shows the basic mechanism they present on their website. The essential idea is pretty straightforward. Take single molecules of DNA, use pyrophosphorolysis to pull off individual nucleotides. The individual nucleotides then get encapsulated in microdroplet reaction vessels where they undergo a reaction which allows then to be detected by a fluorescent signal. My guess would be that the idea is that this might result in longer reads as opposed to Illumina-SBS sequencing. It seems doubtful to me that this process might result in higher accuracy reads.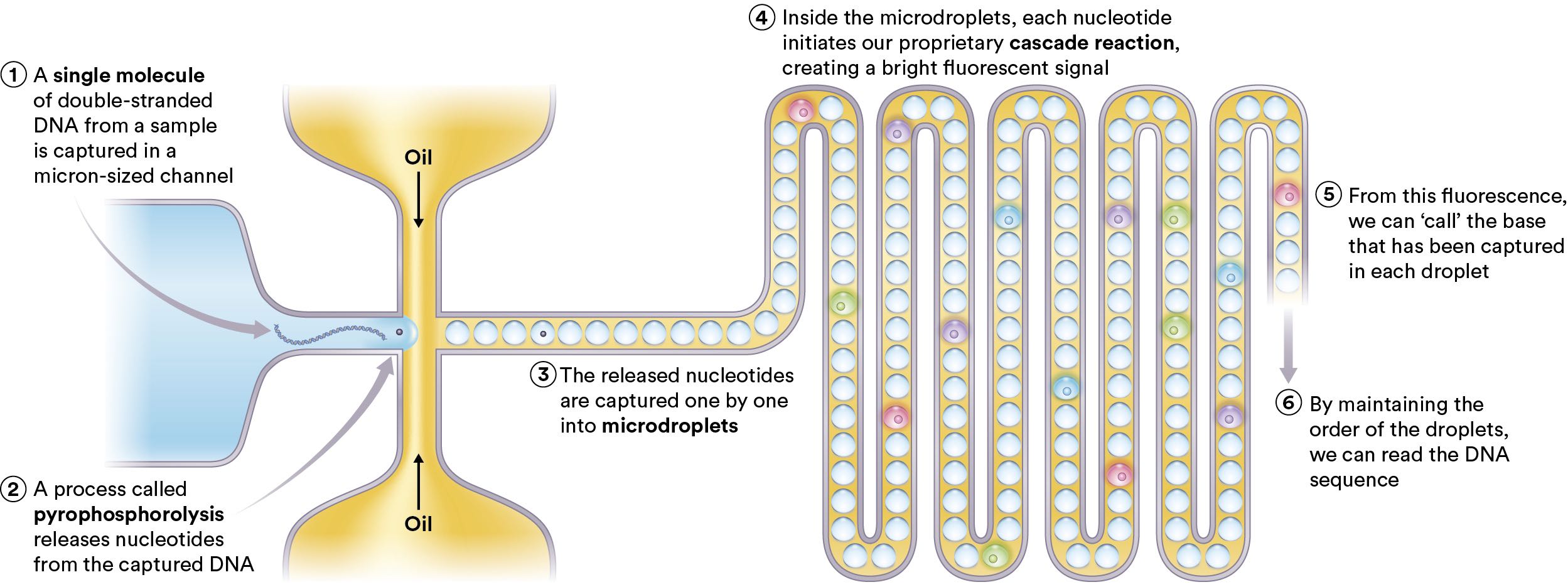 In addition to this method they appear to have patents covering plasmon nanopores, which I’ll cover below. First I’ll cover each part of the process above in a little more detail.
In addition to this method they appear to have patents covering plasmon nanopores, which I’ll cover below. First I’ll cover each part of the process above in a little more detail.
First they need to remove individual nucleotides. The normal way of doing this would be to use an exonuclease. Which is an enzyme that cleaves off bases. Others have proposed using exonucleases for single molecule sequencing (including Oxford Nanopore’s original approach). The patents don’t very clearly state what they’re doing (when are patents ever clear?) but they appear to be using an enzymatic approach, I guess either an exonuclease or a polymerase with some exo activity. The sequencing template is attached to a surface and the nucleotides get chewed off.
They then get stocastically captured in droplets (so there will we a lot of empty droplets, and quite possibly some droplets containing multiple bases). These then get transported through a fluidics system (though a recent patent suggests they might also just be arrayed on a surface [5]).
At this point they have single nucleotides in little reaction vessels, presumably with some reagents. The next stage is therefore to determine which nucleotide is in each droplet. The website currently refers to a cascade reaction and fluorescence, but patents also refer to Plasmon resonance [6].
The cascade reaction appears to work by having the single nucleotide bind to a capture site on an oligo [7]. A substitution-dependent restriction endonuclease comes in and cuts the oligo depending on whether the nucleotide is present or not. The now cut oligo can now be processed by a double-stranded exonuclease. This releases more single bases, hence producing a cascade reaction.
In the process of the above cascade reaction, fluorescent labels are activated. The exact process isn’t described (that I can see) but it could be that a quencher is removed or something similar. Like most patent portfolios, this is just one suggested mechanism… but I’d guess this is the cascade reaction mentioned on the website. That about wraps it up, by detecting the fluorescence (I assume they need multiple labels/lasers) they can call the DNA sequence. There are also patents referring to methylation detection but I’ve not taken the time to read these yet.
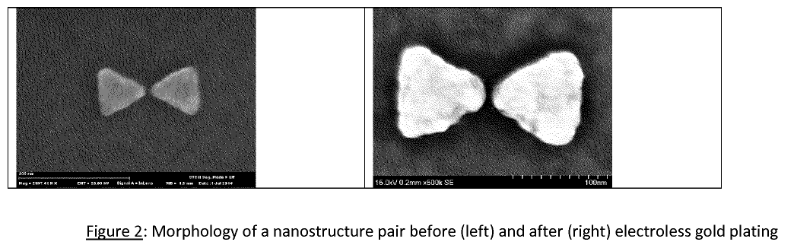
As a final note, a few of their patents refer to plasmon nanopores. This appears to be unrelated to their microdroplet sequencing efforts, and for an early stage company it seemed odd to be experimenting with such a radically different approach. The patents contain SEM images, suggesting that some real work has been done on this system. The approach is obviously also somewhat related to the Armonica approach previously discussed. SEM images below. It seems likely that this may relate to their collaboration wit Hitachi that was reported in 2013 [9].
That about wraps it up for Base4. There’s some interesting tech, and a pile of patents which may no doubt yield further insights. If you’d like to discuss Base4 further (or sequencing in general) leave a comment below or email me (new at sgenomics dot org).
Notes
[1] http://www.base4.co.uk/news/base4-nov17-funding/
[2] Glassdoor reviews (selected mostly negative review containing interesting gossip):
Review 1
Pros
Good people from all disciplines, possible to learn a lot by mixing with people from different backgrounds.
Cons
Micromanagement from senior staff, some of whom have no formal qualifications in science. The CSO left and has not been replaced, leaving scientific oversight in the hands of those less qualified. Very high staff turnover.
Advice to Management
Trust those who you’ve hired, don’t micromanage. There’s no point bringing in postdoctoral level scientists then treating them like technicians.
Review 2
Pros
You can enjoy personal trainer sessions, playing anytime table soccer, free food and drinks.
Cons
The biggest problem is that you need to bare random moody, bully, disrespectful and unprofessional behavior of the CEO who, in addition, has no scientific qualifications/competences.
You might end up to go, everyday, at work with a feeling of terror because there is great probability that the CEO can be mad at you for any reason.
They promised to work in a transparent environment.. but the reality was different.. I discovered bunch of exaggerations and sometimes lies.
The management hires nice and qualified people .. but a great percentage leaves the job after 6-12 months and this slows down the project you work on.
Review 3
Pros
Shiny on the outside. That’s about it. I would not read too much into the veneer of things working smoothly.
Cons
If you like working for a schoolyard bully that has no regard for his employees then go for it.
If you want to do something complete irrelevant to what you were promised before getting the job and if you like to have an adjustment period of about 5 seconds, then again, go for it.
If you like being proud that your are simply stumbling along with no clear vision from the management, then again, fell free to join.
In your shoes I would probably try to work in a different building.
Review 4
Pros
It is a fairly small company with approx. 15 people. It has no strict regulations and you have relatively flexible working time.
Cons
If you are not a DNA specialist or a chemist, do prepare that you need to explain everything from fragment to your manager as they know nothing but they oddly have the confidence that they can recruit people.
Interview Question
After applying online via cv, covering letter and a questionnaire, was asked for Skype interview. One of the most bizarre interviews I’ve ever had. Was asked a single question – ‘why I don’t you have a Ph.D. then?’ to which I replied I’d wanted to get into industry at which the CEO rather abruptly ended the interview.
Was left with the impression that Base4 is very much led by the CEO’s gut instinct and this could be a difficult place to work unless you are a good personality match with him.
[3] https://beta.companieshouse.gov.uk/company/06389614
[4] Interestingly in the funding news in 2017 they state 32 employees. By guess would be that prior to the round there was a hiring freeze, and a few people left.
[5] “Whilst the method described above can be carried out by creating and manipulating a stream of the droplets dispersed for example in an immiscible carrier medium such as silicone oil, we have recently found that the method can advantageously and more effectively performed by printing the droplets directly onto the surface of a substrate as they are formed.” https://patentimages.storage.googleapis.com/15/95/c0/60260db5a928fa/EP3115109A1.pdf
[6] “1. A method for determining the sequence of nucleotide bases in a polynucleotide analyte, the method comprising steps of: (a) generating a stream of droplets at least some of which comprise both (1) a single nucleotide base and (2) colloidal metal particles capable of undergoing plasmon resonance, and (b) irradiating each droplet with electromagnetic radiation to (1) cause the metal particles contained therein to undergo plasmon resonance and (2) the nucleotide base also contained therein to Raman scatter light at one or more wavelengths characteristic of its type. ”
[7] “(a) a first single-stranded oligonucleotide labelled with first and second regions of characteristic detectable element types in an undetectable state located respectively on the X’ and Y’ end sides of a third region comprising a restriction enzyme recognition site element including the capture site and an exonuclease-blocking site on the X’ side thereof (wherein either X’ is 3′ and Y’ is 5′ or X’ is 5′ and Y’ is 3′) and (b) second and third single-stranded oligonucleotides capable of hybridising to complementary regions on the first oligonucleotide flanking the capture site; (2a) either (i) treating the used probe with a conventional or nicking substitution-dependent restriction endonuclease to cut the first oligonucleotide strand at the recognition site if and only if the single nucleotide captured comprises a nucleobase which is substituted or (ii) treating the used probe with a conventional or nicking substitution-sensitive restriction endonuclease to cut the first oligonucleotide strand at the recognition site if and only if the single nucleotide captured comprises a nucleobase which is unsubstituted; (3) digesting the first oligonucleotide strand of the used probe with an enzyme having double-stranded exonucleolytic activity in the X’-Y’ direction corresponding to the first oligonucleotide to yield detectable elements derived from either the first region, the second region, or the first and second regions in a detectable state and a single-stranded fourth oligonucleotide which is at least in part the sequence complement of the first oligonucleotide; (4) reacting the fourth oligonucleotide with another first oligonucleotide to produce a substantially double-stranded oligonucleotide product corresponding to the used probe; (5) repeating steps (2a), (3) and (4) in a cycle and (6) detecting the detectable elements released in each iteration of step (3) wherein if the endonuclease employed is of the conventional type the second or third oligonucleotide includes an endonucleolysis-directing linkage at or close to its X’ or Y’ end respectively.”
[8] “Further information about the pyrophosphorolysis reaction as applied to the degradation of polynucleotides can be found for example in J. Biol. Chem. 244 (1969) pp. 3019-3028. The enzyme which is preferably employed in this pyrophosphorolysis reaction is suitably selected from the group consisting of those polymerases which show essentially neither exo- nor endonuclease activity under the reaction conditions. Examples of polymerases which can be advantageously used include, but are not limited to, the prokaryotic pol 1 enzymes or enzyme derivatives obtained from bacteria such as Escherichia coli (e.g. Klenow fragment polymerase), Thermus aquaticus (e.g. Taq Pol) and Bacillus stearothermophilus, Bacillus caldovelox and Bacillus caldotenax. Suitably, the pyrophosphorolytic degradation is carried out in the presence of a medium which further comprises pyrophosphate anion and magnesium cations; preferably in millimolar concentrations. ” https://patents.google.com/patent/WO2014167323A1/en?oq=WO2014167323+
[9] https://www.genomeweb.com/sequencing/base4-hitachi-developing-single-molecule-nanopore-based-sequencer
“Privately owned Base4, the partners said, has developed a method of increasing the signal from the single molecule passing through the pore by using laser light enhanced by gold structures. The technology, they added, allows the signal to be read from unlabeled DNA, minimizing sample preparation.”