AxBio – Nanopore Sequencing Startup
This post previously appeared on the substack.
AxBio was founded in 2016 by Hui Tian and Igor Ivanov. Hui was previously Vice President of Genia Technologies. The company appears to be funded by Tsingyuan (now Foothill) Ventures, who interestingly also invested in Quantapore.
AxBio appear to be developing a “forth generation” DNA sequencing platform, which they say can “can shorten the human genome sequencing from the current 1-2 weeks to one day”.
In addition to this, they (surprisingly to me) also sell a number of generic extraction kits:
It’s hard to find out much about AxBio, but they suggest they have a 1 million sensor chip. Other reports suggest this is a “nanopore sequencing by DNA synthesis”..approach using a…“hydrodynamic chip combining a silicon CMOS base and a biological membrane”… “together with a polymerase and labeled nucleotides”.
This kind of suggests an approach similar to Genia, where tagged nucleotides are incorporated and read in-situ:
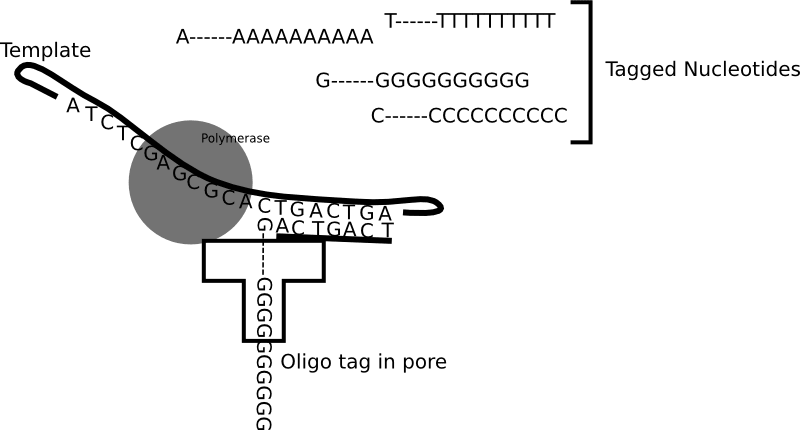
Unfortunately, I’ve not seen much from Genia/Roche since I last wrote about them in 2016. Suggesting progressing the platform to a point where it’s competitive is proving problematic.
AxBio’s patents suggest a similar approach, where a polymerase is used in-situ to synthesis a strand containing labeled nucleotides.
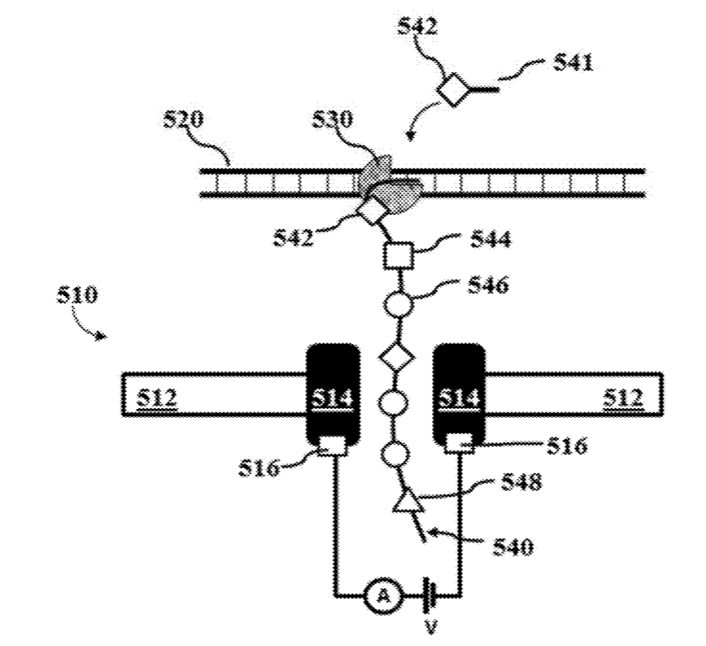
Synthesizing in-situ may help slow the translocation of the strand through the pore, which would generally be required for any ionic current detection method. They also seem to suggest reading the same strand multiple times, using rolling circle amplification to create multiple copies of the input DNA.
This nanopore-SBS approach has the obvious disadvantage that you are not sequencing the original material and as such will not see base modifications, or be able to natively sequence RNA.
As well as protein nanopores, Axbio patents discuss solid state nanopores at length which suggests a different sensing mechanism is also being considered. In particular the patent states that impedance sensing may be used to sequence by “detect one or more signals indicative of an impedance or impedance change in the nanopore when the at least portion of the tag is within the nanopore”.
This sounds pretty ambitious. A single tag would need to create an impedance change within the volume of the nanopore which can be detected during the short period of time it resides within the pore. In addition to this, you will likely have impedance contributions from multiple tags, background impedance contributions from the buffer, and potentially if the electrodes a close enough, tunneling currents between the electrodes.
The patents don’t contain any example procedures or datasets that I could see. So I went looking for support for impedance sensing approaches in the literature. My friend Shohei manage to find a publication where a single molecule induces a capacitance change by displacing a membrane. But this is a big leap from detecting impedance changes created by single molecules, and we were unable to find much support in the literature for this sensing method.
Outside of nanopores, scanning capacitance microscopy is a relatively established probe microscopy technique. In this article exploring the limits of SCM, they suggest that best resolution you can obtain in the Z direction is 1nm, and 5nm in XY. Here using a bandwidth of ~1KHz. For the most part SCM appears to be used for imaging semiconductors and similar planar surfaces. This is clearly a lot easier than measuring single molecule tags.
5nm would suggest we would see contributions from ~15 bases/tags. Which would be significantly worse than the ~5 you see with protein nanopores.
So overall, the impedance based approach seems like it would be really hard to get working.
AxBio however appear to suggest that they have “released the latest single-molecule gene sequencer” so it will be interesting to see what actually appears. They talk about a 1 million sensor thumb nail sized chip. This also suggests a solid state approach, as achieving this density with protein nanopores is likely challenging. So, I remain curious as to what will actually appear and will be keeping an eye out for any data releases from AxBio.
Disclaimer: As always, you should be aware that I have equity in sequencing companies (based on prior employment) and am current working on a novel sequencing approach.