Universal Sequencing Technology
Another in my ocassional series on currently active DNA sequencing companies. Today, Universal Sequencing Technology.
Business
There’s not much information on Universal Sequencing Technology. The Delaware state database lists their incorporation date as 9/1/2016. However their international patent has a priority date of 30th October 2015. Two employees are lists on LinkedIn (Robert Roorda, Head of Instrumentation and Scott Kozak, Chief Business Officer).
I’ve not been able to find any information on funding.
Technology
There is one patent assigned to UST [1].
This concerns itself almost exclusively with control the motion of DNA in nanopore sequencing platforms. One of the major issues with nanopore sequencing is slowing the passage of the DNA through the pore so it can be read. Without being slowed, the DNA will pass through the pore at millions of bases per second.
The patent doesn’t limit itself, or even focus on, any particular type of nanopore (protein or solid-state). Nor do they discuss Ionic current blockage versus tunnelling current detection in any great detail.
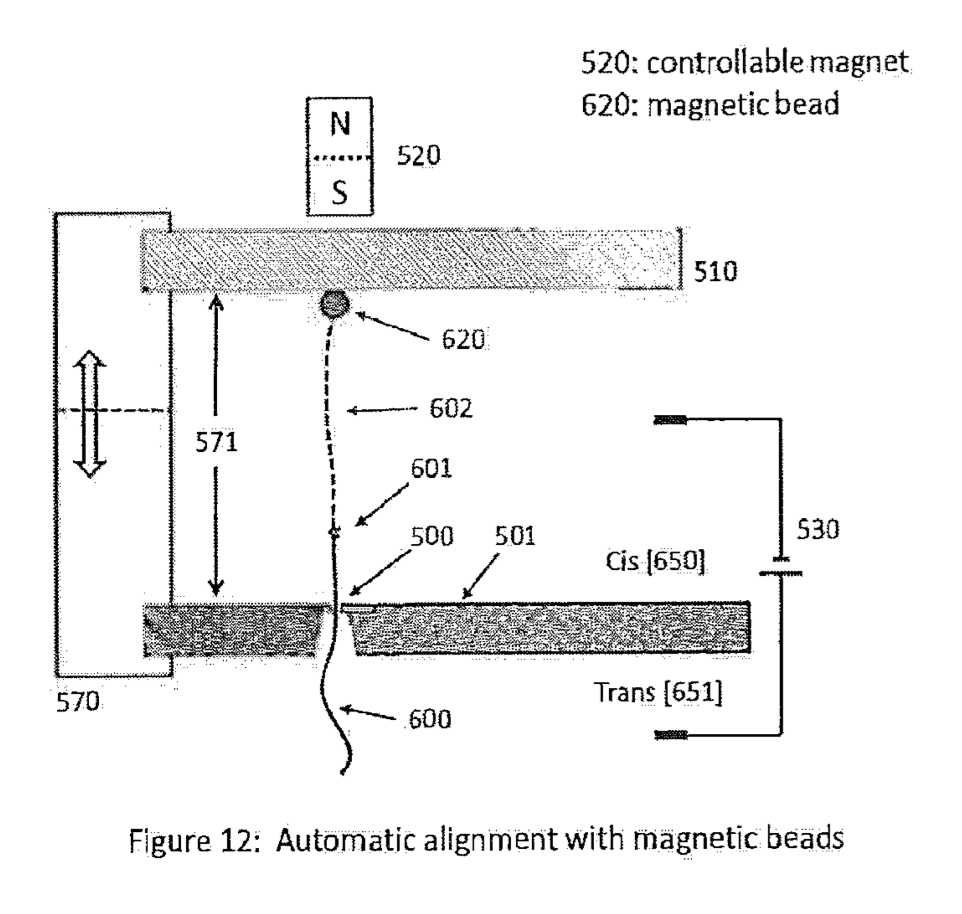
The crux of the application is really the use of non-enzymatic methods to control the motion of the DNA. A strand to be sequenced is attached to a substrate of some kind (for example a bead). The strand is then dragged into the pore (under a bias voltage). However because the DNA is attached to a substrate on the other end, it can’t get pulled all the way through the pore.
The substrate is then moved, such that different parts of the strand sit in the pore. This is not unlikely some of the work of the Keyser lab where optical tweezers are used to control the motion of a bead to which a strand of DNA is attached.
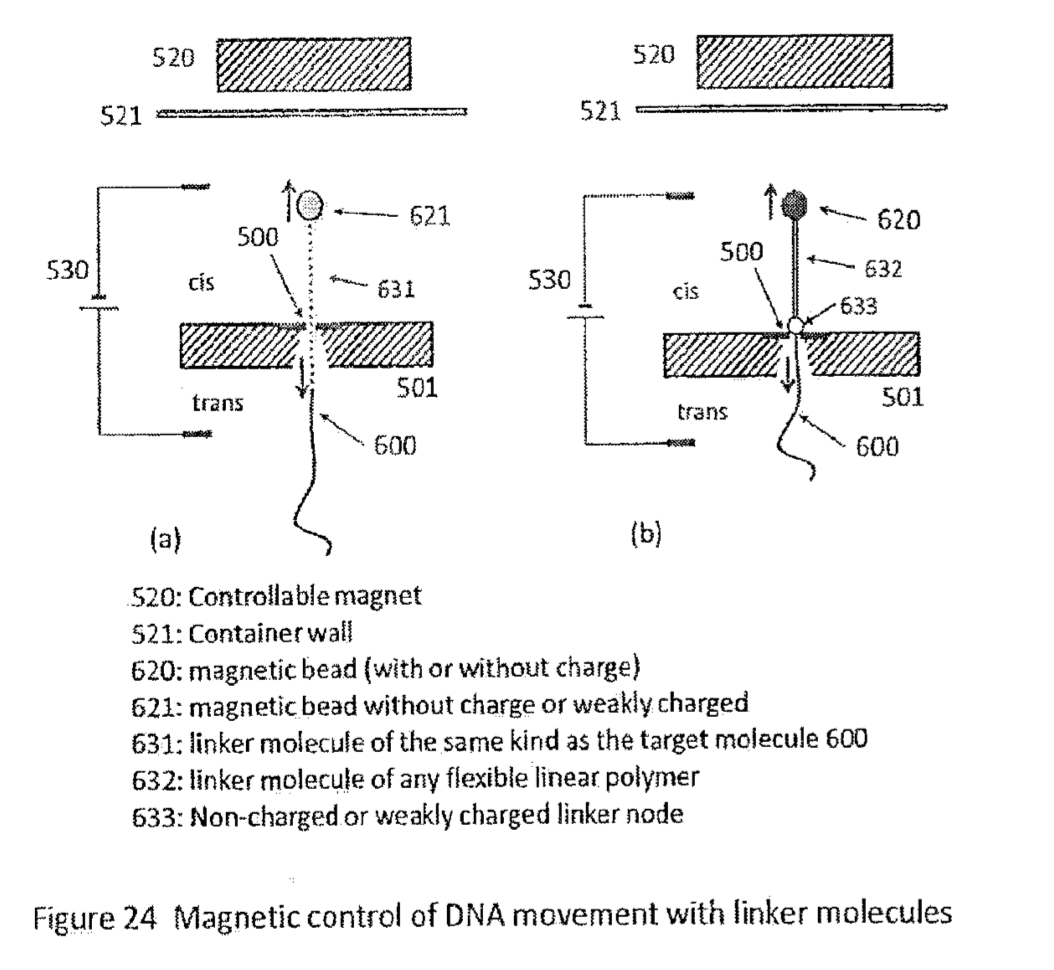
There are a number of ways to control motion with the nanometer to sub-nanometer precision required. Many of these have been developed for use in scanning probe microscopy. There are a few methods proposed in the patent, but the preferred scheme seems to be the use of a sequencing strand attached to a magnetic bead:
The bead itself in uncharged (in some embodiments at least). It gets pulled through the pore by virtue of the of the charge on the DNA strand. They can detect when the strand enters the pore, and at that point setup a magnetic field to balance the force on the strand. They can then increase or decrease the magnetic field strength to move the strand in and out of the pore (this has been called “flossing” elsewhere). The strand can sit, ideally relatively static, in the pore while the signal from a single position is read.
As usual there are a number of modification to the basic scheme proposed, longer linkers… spacing strands etc:
The idea is pretty reasonable, and through the use of optical tweezers (and potentially other methods) has obviously been tested in academic work. This patent doesn’t show any data, but I’ll be interested to see how the approach plays out in practice.
Notes
[1] https://patents.google.com/patent/WO2017075620A1