Dreampore – Protein Sizing
This post was originally published on substack.
I came across another patent from the Dreampore folks when investigating the platform, and thought it might be interesting to write up. It’s actually not clear if this patent has been licensed to Dreampore, but 3 of the inventors are listed as working at Dreampore so it seems reasonably likely.
The patent appears to be based around work from a 2018 paper. However the patent itself is interesting to me in part because of the way it’s written and the scope of the claims.
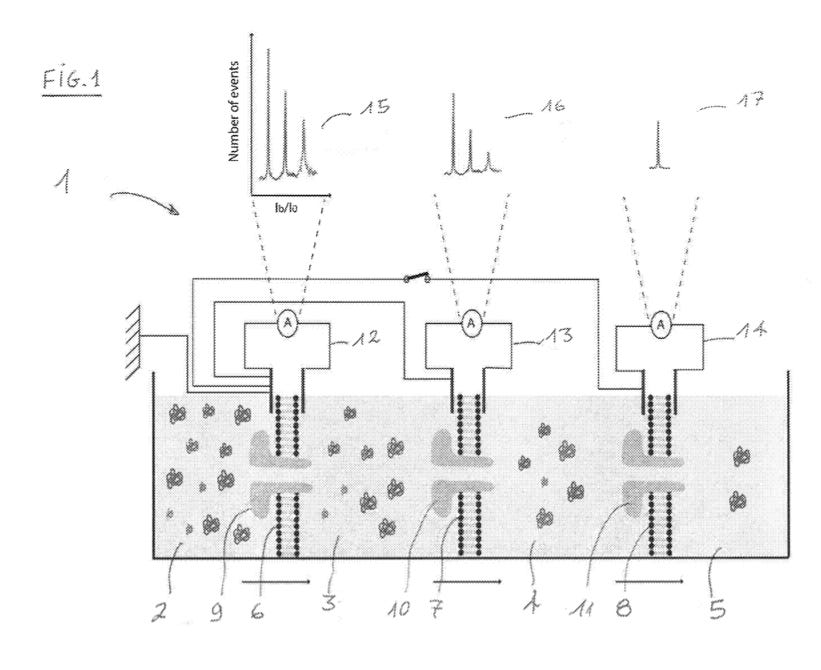
The patent is framed around identifying impurities (protein fragments) in a 96% pure sample. They state that “it is impossible to identify these fragments by classical mass spectrometry or by HPLC”. As in their publications they use aerolysin nanopores, a three chamber device is presented:
It’s not clear to me why you want to use three chambers. They suggest that pure product could be removed from the second chamber… but don’t describe how or why this is useful. They also don’t appear to use a three chamber device in the paper.
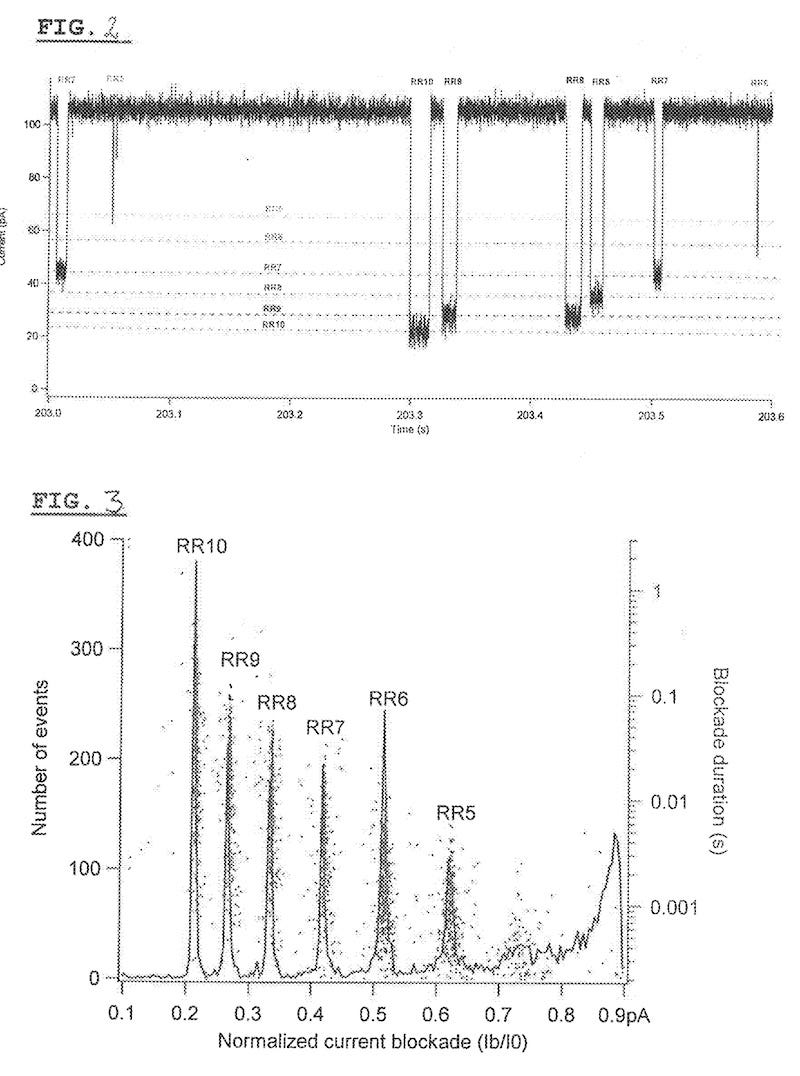
They show what appears to be experimental data from translocations of a protein (RR10). While it’s not stated in the patent, it seems clear from the publication this is a 10 amino acid long arginine homopeptide (RRRRRRRRRR). What they shown is that they and distinguish between homopeptides of lengths between 10 and 5 amino acids.
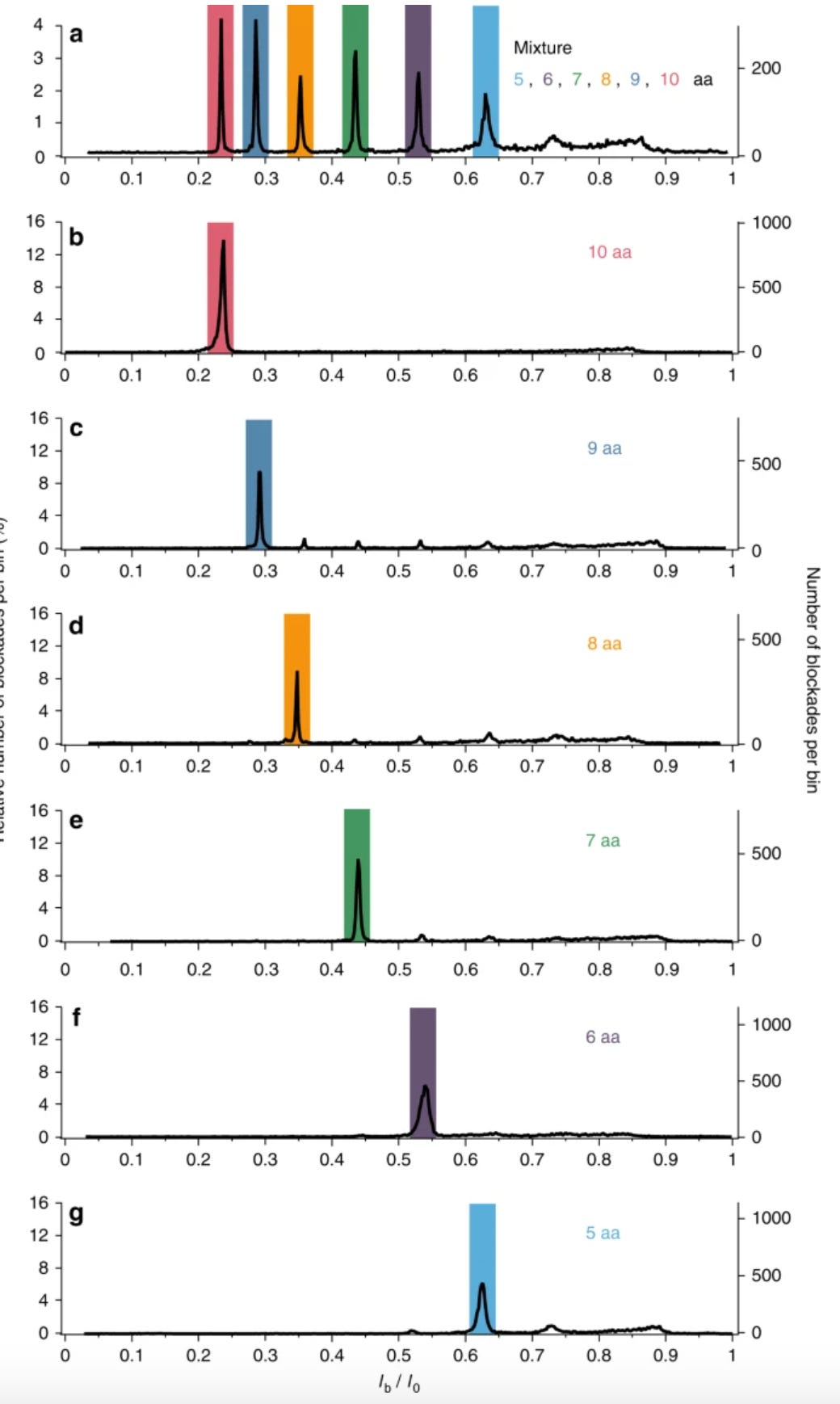
The paper further clarifies this showing samples containing single peptide types, as compared to the mixture:
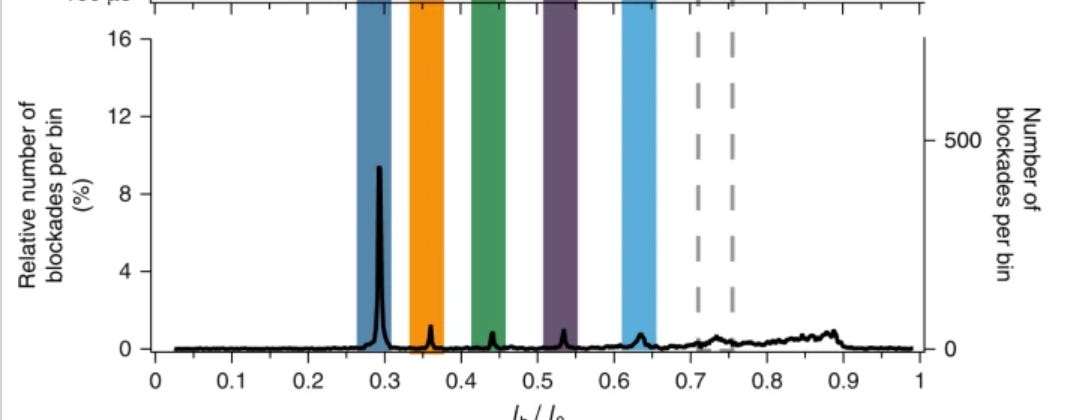
They then take this further, and show relative concentrations of peptides in a sample from a supplier claiming 98% purity:
So, overall they show size determination at a the single amino acid level and present an application in the classification of impurities during reagent production. As far as this goes, this is fine.
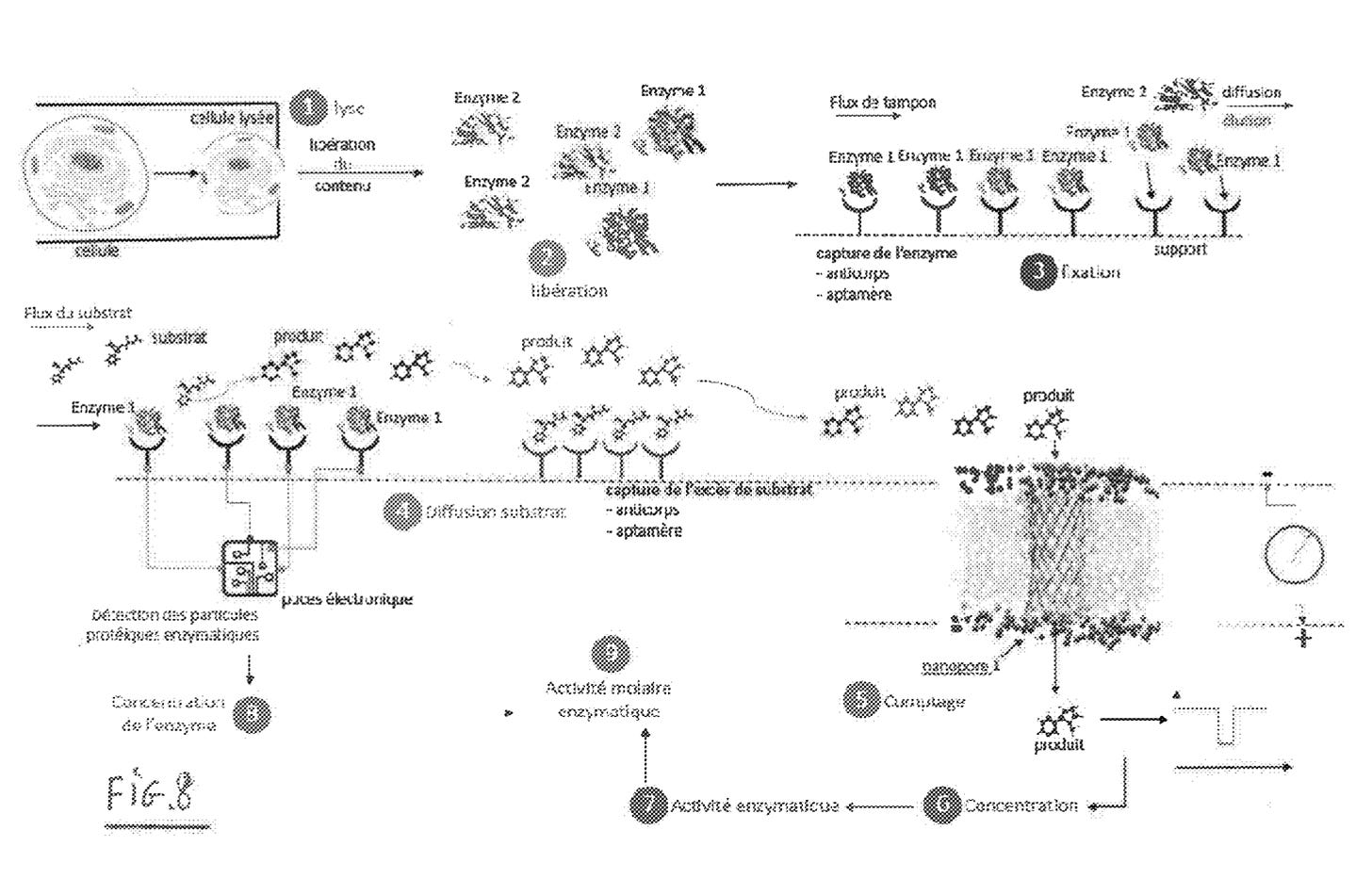
The patent then describes how this might be used to measure enzymatic activity. The idea is to quantify enzymatic activity on the single cell level. Enzymes are captured on a solid support. You then flow in the enzymes substrate and pass the product through a nanopore. This seems like an interesting research problem, but I’m less clear on the market for such a device, and there’s no experimental data.
The patent overall seems quite weak (I don’t blame the authors for this, it seems like it might have been rushed).
The single independent claim in this patent reads:
“The use of an aerolysin nanopore or a nanotube for the electrical detection of peptides, protein separated by at least one amino acid and other macromolecules such as polysaccharides or synthetic or natural polymers present in a preparation where said nanopore or nanotube is inserted into a lipid membrane which is subjected to a difference in potential of over −160 mV, in a reaction medium comprising an alkali metal halide electrolyte solution with a concentration of less than 6M and at a temperature of less than 40° C., and where said use is intended to differentiate said peptides, proteins and other molecules according to their length and their mass.”
Which seems very narrow. And what does over -160mV mean? If I look to the paper I can understand that they term -100mV as greater than -50mV, but it’s obviously not in terms of absolute magnitude. And shouldn’t this be specified in relation of the cis/trans side of the pore? The specification doesn’t provide much clarity here.
There are numerous errors in the patent for example confusing voltage and current (“voltage of 25pA”). And other statements and typos that make me think the patent was rushed. Which seems like a shame…
In terms of the technical approach itself, I’m not sure if measuring reagent impurities is a compelling market. But being able to detect single amino acid differences seems like a step toward some more interesting applications.