BGI – Miscellany
I’ve previous discussed the BGI Business, and the DNA sequencing technology they acquired from Complete Genomics. In this post I want to tie up a few loose ends. In particular, I was curious to see if the BGIs patents covered any other approaches. From what I can tell, their patents don’t cover other approaches in much depth, but there are a few interesting bits and bobs!
MGI Tech
MGI Technologies is a wholly owned subsidiary of the BGI Group. They’re currently looking to IPO on the Hong Kong stock exchange, and are attempting to raise 1B USD [2]. They have a number of patents assigned to them, these mostly concern themselves with sample prep methods. However there is one patent on DNA sequencer chip design [3] it looks very much like the BGISEQ50 chip shown on one of their sites:
SBS-patent?
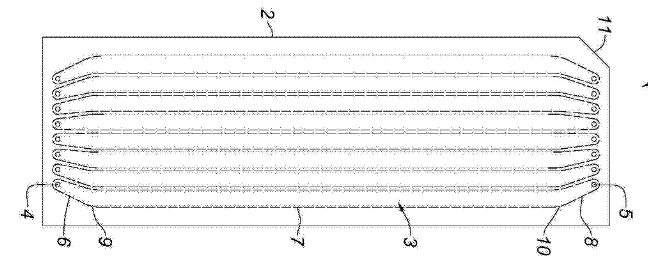
There’s a patent from the BGI which seems a bit odd to me [5]. It essentially seems to cover a flowcell and SBS sequencing methodology that looks very much like the Illumina process. In fact, the flowcells look almost identical:
It’s possible that there’s something I’m missing here, because the patent is in Chinese, and the translation isn’t great. But it’s interesting none the less. The patent doesn’t seem to cover specifics of the chemistry or imaging system, but seems to talk about the overall approach. MGI Tech also seem to have a patent covering a 2 color optical system [7], not dissimilar to the Illumina 2 color sequencing approach.
A number of other patents cover optical systems for DNA sequencers, but don’t appear to discuss a particular sequencing approach [6].
Nanopore Patent
There’s one nanopore patent [1] assigned to the BGI Shenzhen. It mostly concerns itself with a “multi-pass sequencing” approach, to reduce error rates. In this approach a “concatemer nucleic acid molecule comprising a plurality of copies of the target sequence” is used to reduce the overall error rate.
The patent also discusses “membranes of randomly distributed nanopores” as opposed to ordered arrays used in most systems. They suggest that nanopores are instead randomly distributed on a single membrane. Optionally they can be cross-linked to fix them in place. Micro-pipettes are then are then used to create a seal around one side of the nanopore. This is similar to a traditional patch-clamp approach, and I don’t quite understand what they’re getting at here. It seems like they suggesting billions of pipettes are used. This seems like a problematic system to build.
Notes
[1] Nanopore methods patent http://www.freepatentsonline.com/WO2016077313A1.pdf
[2] https://www.bloomberg.com/news/articles/2018-04-25/china-dna-giant-s-equipment-arm-said-to-seek-1-billion-funding
[3] Chip design: http://www.freepatentsonline.com/WO2018081920A1.pdf
[4] https://www.utsouthwestern.edu/labs/next-generation-sequencing-core/sequencing/
[5] https://patentimages.storage.googleapis.com/9d/a9/9c/697f0ed8d84cab/CN205133580U.pdf another patent, https://patentimages.storage.googleapis.com/8a/13/0b/727f18776a8f2e/CN205368331U.pdf also describes a 2 lane flowcell.
[6] https://patents.google.com/patent/CN205368376U
[7] https://patents.google.com/patent/CN206607236U