Business
Centrillion was founded in 2009 by ex-Affymetrix staff. Of the current leadership team only Wei Zhou (CEO) was at Centrillion at its inception and previously worked at Affymetrix. Wei Zhou worked at Affy as an SVP and left after 8 years in 2008 according to LinkedIn [1].
Centrillion has a more complex corporate structure than I’m used to seeing in a startup. The governing entity for Centrillion appears to be Centrillion Technology Holdings Corporation which is a Grand Cayman corporation [9]. Centrillion Biosciences Inc. [2] is a subsidiary of the holding company, and is located in Mountain View.
They appear to have offices in Palo Alto, Portland, Hangzhou-China and Hsinchu-Taiwan [3]. They mention that they acquired “assets” from Perlegen and Affymetrix, but it’s not very clear what these assets are. Given that they also tried to acquired Affymetrix in 2016 I would guess that there was IP and staff and other assets that they were interested in acquiring at that point (this brought another company into play, Origin Technologies Corporation, LLC) [5]. Affy did not accept the Origin offer, suggesting that the ThermoFisher bid was more realistic [8].
Centrillion seems to be funded mainly by Chinese investors. SummitView Captial [4] is among these, and also backed their Affymetrix bid.
Centrillion has launched at least one service, Tribecode. This seems to be a genetic genealogy testing service. Reports suggest is uses low coverage Illumina sequencing. At present the tribecode website (www.tribecode.com) points to an unconfigured webserver. The service is however mentioned on the Centrillion website. It’s unclear if this is a temporary issue (it’s been offline for at least a couple of days) or if the service is no longer available.
Glassdoor reviews for Centrillion are interesting [17], many reviews complain of cameras monitoring staff work areas and being forced to clock in and out of the office (and reprimands for being in the office less than 8 hours). One review notes “Company appears to be a front for sheltering Chinese capital investors rather than an actual product or technology driven enterprise.”. I tend to take glassdoor reviews with a grain of salt, but it’s interesting gossip.
Last week (19th July 2018) Centrillion are reported as raising 58.78M USD [10] [11]. In total they appear to have raised in excess of 86.79M USD [11]. In addition to this, they appear to have received 650,000USD in SBIR grant funding [12].
Technology
The Centrillion website has a services link, this currently points to services.centrillionbio.com which I can not access currently (timeout). Looking at archived copies of the site at the Internet archive suggests that all services are carried out on Illumina instruments. In particular they reference Hiseq2000 series instruments (which are now quite old): “sequencing to desired coverage using Illumina HiSeq2000 or HiSeq2500 instrument”.
Centrillion also runs “Molecular Vision Lab”. This subsidiary seems to have a number of panels available for vision related genetic disordered. The sites states “This new test is a capture based panel with baits designed and manufactured by our parent company, Centrillion Technologies.” [13].
None of these services sound quite like the statements from Centrillion’s factsheet which suggest development of a DNA sequencing platform:
“Centrillion is leveraging an extensive intellectual property portfolio and the latest advances in engineering, chemistry and biochemistry to develop an array of solutions, from targeted sequencing to chromosome scale sequencing; from genotyping to 3D gene expression, all based upon a unifying genomic analysis technology.
Centrillion’s work is supported by a new generation of sequencing-compatible DNA chips, which combine the high density of first-generation chips with the high-quality probes of more recent innovations, all in one affordable and scalable mode.” [16]
So I’ve tried digging a bit further to find more details of sequencing technologies they might be building.
One patent, “Methods and Systems for Sequencing Long Nucleic Acids” [14], discusses a modification of SBS protocols to enable longer read sequencing. The method relies on the use of existing SBS IP and discusses demonstrations on Illumina and Ion Torrent platforms. Essentially they suggest performing multiple reads against the same clusters, at offset positions.
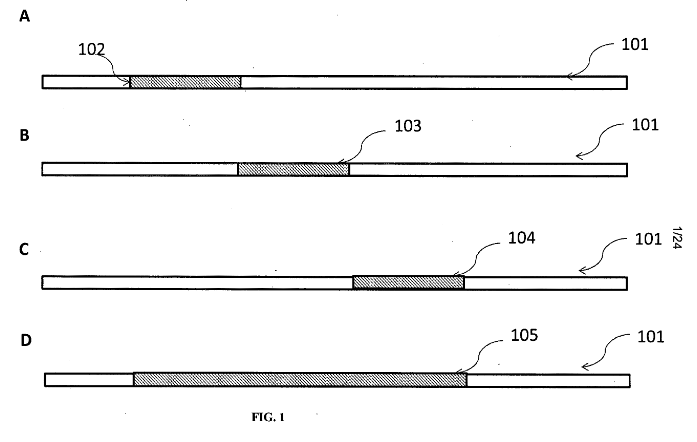
So you would perform a normal single sequencing read. Then remove the synthesized strand. Then re-prime the clusters. This essentially resets the sequencing experiment. You then use one of a number of methods to advance the start position. A few methods are discussed. Most of these seem to rely on what they call “limited extension”, where they incorporate bases but limit the number incorporated somehow e.g. by
* Limiting the amount of time.
* Leaving out one base, so the extension stops at that base.
* Having one base use a reversible terminator to stop the extension.
The examples they present use what they call +S, dark base extension. Dark bases are just native nucleotides, i.e. unlabeled. On an Illumina instrument after “resetting” the clusters they perform a number of cycles with mixtures of these dark bases. For example you might first do a cycle containing A,T and G but not C. This would advance the cluster to the next C position. Then you wash, and do another cycle with C,A,T but no G. A number of these cycles can be performed to advance the read position. After you’ve completed this “limited extension” process you can perform your read as normal.
Neat idea, there’s some data in the patent but I haven’t taken the time to dig into it.
There’s another patent “Native Extension Parallel Sequencing” [15] which also seems to use the limited extension concept. In this patent, a capture system is used. Using this you can have multiple spots/regions which contain the same template. The patent then uses a system which to be frank I don’t quite understand, to obtain partial information from each spot which is assembled into a complete read. It seems interesting, and if I had time I’d like to dig into this patent more and better understand it.
These are both quite old patents… more recently a paper co-authored by Centrillion staff has been published. This paper is discusses on surface library construction. Again this looks interesting, but I’m more interested in sequencing technologies.
There are a few other bits and bobs like this patent for a reversible terminator. But nothing that indicates to me serious development of an SBS platform. So I’m still curious as to what they are actually developing. It seems possible that there interests have shifted from sequencing toward prep/capture/library construction and running a services business.
Notes
[1] Leadership team from the Centrillion factsheet, who worked at Affy. Durations and dates from LinkedIn:
Wei Zhou (CEO) was SVP at Affymetrix until 2008 (>8 years)
Janet Warrington, VP R&D until 2008 (12 years). Joined Centrillion 2014.
Glenn McGall, VP; Research and Development, Chemistry until 2012 (5 years)
Suzanne Dee, Director, Expression Product Development, Jul 2014 (>18 years)
[2] https://www.bloomberg.com/research/stocks/private/snapshot.asp?privcapId=72011751
[3] Office locations from job posting, https://startup.jobs/62911-research-scientist-at-centrillion
“We have advanced research laboratories in Palo Alto, CA; Portland, OR; Hangzhou, China and Hsinchu, Taiwan”
[4] http://en.summitviewcapital.com/plus/list.php?tid=10
SummitView Capital backed acquisition: https://cen.acs.org/articles/94/i13/Former-employees-seek-acquire-Affymetrix.html
[5] https://www.sec.gov/Archives/edgar/data/913077/000161577416004576/s102858_dfan14a.htm
Mar. 27, 2016: https://seekingalpha.com/article/3961150-week-review-origin-centrillion-offers-1_5-billion-affymetrix
Centrillion fail to buy Affymetrix: https://cen.acs.org/articles/94/i14/Thermo-Fisher-wins-contest-Affymetrix.html
[6] Tribecode review: https://thegeneticgenealogist.com/2015/08/01/a-review-of-tribecode-by-centrillion-biosciences/
[7] Funding rounds:
https://www.crunchbase.com/organization/centrillion-biosciences
[8] https://www.genomeweb.com/business-news/affymetrix-thermo-fisher-rebuff-origin-technologies-purchase-offer
Key quotes:
In reviewing the proposal, Affy said that it has concluded that Origin appears to be a “newly formed shell entity with no assets of which Affymetrix is aware, and whose sole source of funding for the proposed transaction is $1.5 billion in potential debt commitments.” Moreover, this $1.5 billion is not enough, according to Affy, to cover the proposed deal in full.
“The proposal put forth to Affymetrix by Origin Technologies, a newly created shell entity relying on a vague and insufficient financing package from a Chinese firm, is highly uncertain and speculative and does not constitute, and could not reasonably be expected to lead to, a superior proposal under the merger agreement, and Affymetrix and its board of directors could not reasonably determine otherwise,” Marc Casper, president and CEO of Thermo Fisher, said in a statement.
[9] Patents list the Centrillion address as:
Centrillion Technology Holdings Corporation
Maples Corporate Services Limited
PO Box 309, Ugland House
Grand Cayman, KY1-1104(KY
UNITED KINGDOM
Click to access Patent_18052018.pdf
[10] Funding history: https://www.whoisraisingmoney.com/centrillion-technology-holdings-corp
[11] http://www.xdata.co/story.php?a=000147383518000004&c=1473835
“Centrillion Biosciences received two commitments for its $31.59 million financing round. Investors committed to buy 89% or $28.01 million worth of equity three weeks ago. Based on the offering’s structure, the company has until June 2019 to raise an extra $3.58 million. A total of five unregistered securities offerings closed by the company raised an estimated $58.78 million.”
[12] https://www.sbir.gov/sbirsearch/detail/700133
[13] https://www.molecularvisionlab.com/mvl-vision-panel/
[14] http://www.freepatentsonline.com/WO2012134602A2.pdf (2012)
[15] https://patentimages.storage.googleapis.com/d2/b9/a5/3aea1e5c2405b8/US20120083417A1.pdf
[16] http://www.centrilliontech.com/wp-content/uploads/Centrillion_FactSheet.pdf
[17]
Selected Glassdoor reviews:
1:
Cons
Every con mentioned here is true (I advise only looking at the one star reviews for an accurate portrayal of the work environment).
Advice to Management
At a minimum, fix what is mentioned in these reviews. Management and HR need to be addressed and your employees are miserable. Stop trying to get your current employees to write reviews on this app to raise the company ratings. Instead of trying to deceive future applicants, redirect your efforts to focus on your current employees’ well being.
2:
I worked at Centrillion Biosciences full-time
Pros
Some people on the exec team are the best mentors you can have. They are encouraging and do care deeply for the employees. At times they have tried their best to shield us from the absurdity of the other half of the exec team.
Cons
There are so many cons about this company that many have already touched on in past reviews. I will point out the two below that is most concerning to me.
I worked at Centrillion for 1.5 year before leaving for a better opportunity. Since my departure, I have heard some things have gotten better where others have declined. Centrillion is a very secretive company with absurd company polices. One of the craziest company policy is clocking in and out as mention in other reviews. Everyone at the company except the interns are all on salary but it is required that you clock in and out everyday for 8 hours each day. If you are not physically in the office for 8 hours, you will be reprimanded by HR.
Starting salary is low — starting salary for research assistant can be as low as $50,000. There are ‘yearly’ raises but again it is barely enough to live on in the Bay Area or enough to keep up with the standard.
Advice to Management
Management does not listen or care as long as they can save a quick dollar.
3:
I have been working at Centrillion Biosciences full-time (More than a year)
Pros
My paycheck never bounced…
Free lunch – but who doesn’t have that?
Cons
Poor communications
Paranoid management
Managers unclear about roles
Teams not sync’d up
Operations and HR are unprofessional
Any positive reviews you see are posted by the company
Advice to Management
Move to China where you spend your time anyways
4:
Pros
– Most of the people are smart and easy to work with.
– Some of the senior management tries.
– $200 towards lunches.
Cons
– Many people are unhappy and there is an air of negativity creating a lack of motivation or enthusiasm.
– CEO micromanages everyone and pins employees against each other making it highly competitive among members of the same team.
– Secrecy is highly encouraged.
– Many are not sure why they are working on certain projects.
– There is one woman managing operations, facilities and purchasing.
– Level of education plays a major roll in deciding title, pay raise and promotions- not level of experience.
– Perks are lacking compared to other companies in the area.
– There are internal cameras pointing towards employee seating area.
– Full time employees must work a full 8 hours on a daily basis excluding any breaks taken or lunch.
– Average amount of time an employee stays with the company: 6 months -1 year.
– Management forces you to take few days of employee earned PTO during christmas break every year.
– HR creating false reviews on glassdoor to boost the overall rating!Show Less
Advice to Management
Treat your employees well if you’d like to retain them.
5:
I worked at Centrillion Biosciences full-time (More than a year)
Pros
-Some of the employees here are okay to work with
-Catered lunches but only about 80% of the month
Cons
-Retention rate of employees is discouragingly low (<1 year) and no concern is shown by management
-Very little opportunity for growth (again, management does not seem to care or prioritize their talent)
-Compensation and benefits below industry standard. The company is stingy with compensation.
-Unprofessional Operations group that was also acting as an HR group (concerns and questions are either ignored, delayed, or show lack of interest)
-The way the company is set up creates a paranoid atmosphere in the company (surveillance, clocking in, internal discussions discouraged, feels like you’re being monitored)
Advice to Management
-Invest in employee’s growth and allow them to have competitive wages to afford Silicon Valley housing
-Executive group needs to be more transparent with empolyees
6:
I worked at Centrillion Biosciences full-time (More than a year)
Pros
There are some very good people working there, and the funding appears to be adequate to make payroll. Annual raises for most employees. Free catered lunches.
Cons
Company appears to be a front for sheltering Chinese capital investors rather than an actual product or technology driven enterprise. Management is not interested in employee retention or development. Average employee duration is about one year. Essentially no path to advancement, a secretive and manipulative executive group operating without clear principles or ethics. Compensation and benefits are sharply below the local industry standard. Non-scientific sweatshop atmosphere, junky and poorly organized work areas, internal discussions are discouraged, surveillance cameras pointed inward, rank and file employees are required to clock in and out to leave the building.
Advice to Management
Stop trying to hide what you are from employees, nobody who has worked there more than 3 months believes what you say. Take some business psychology courses.
7:
I worked at Centrillion Biosciences (Less than a year)
Pros
Some good people to work with
Decent place to gain some work experience
Lunch provided by company although that is a norm in Bay Area startup
Cons
Horrible company culture
The worst HR policies. There is absolutely no concept of work life balance. Everyone is required to clock in and out. The management tells the employees that it is for “safety” but the HR keeps tabs on work hours. Workers are required to be at site for 9 hours and anything less leads to censure
Very secretive culture spread by the upper management. No one knows what the other is working on or even what the product is.
Advice to Management
Your employees are your peers not your servants. Treat them with respect.