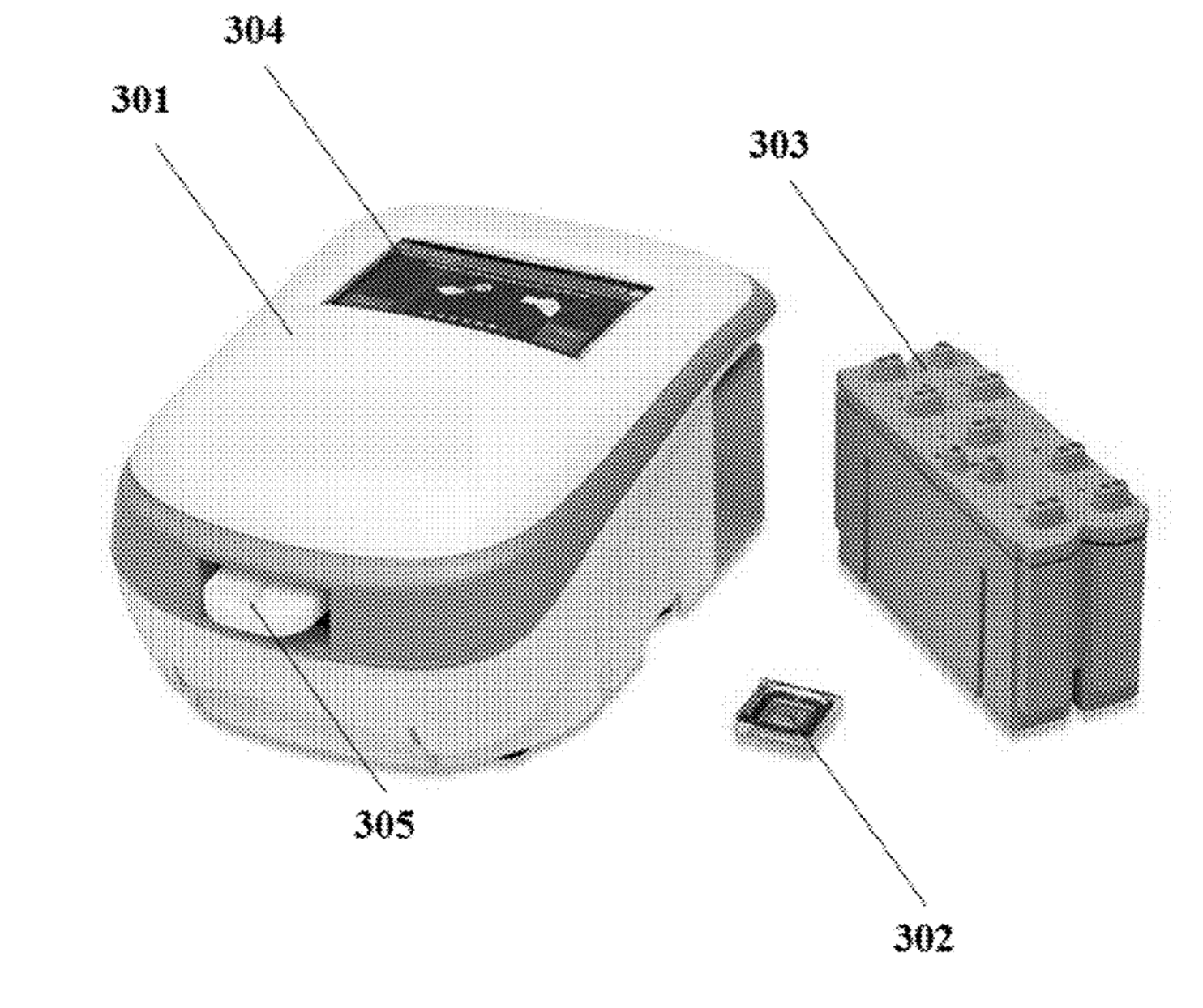
Sequencing system image from patent.
Building on my list of sequencing companies. I’ve put together a few brief notes on Genapsys.
Business
Genapsys was founded in 2010. They have raised in excess of 84M USD in total (110M USD according to their website, 84M accounted for on crunchbase). Investors appear to include Decheng Capital, IPV Capital, Plug and Play Ventures and possibly Ampersand Capital Partners. Yuri Borisovich Milner (DST Global) is also said to be an investor. Their last round was on January 2018, where they raised 32.5M USD, Series C. In addition to venture funding they have received some grant funding (~4M USD) [1].
The company was founded by Dr. Hesaam Esfandyarpour and incubated at the Stanford Genome Technology Center.
Technology
The patents I’ve reviewed describe two key components to the Genapsys approach. The first is a method for confining DNA and reagents using Virtual Wells [3]. The second is a method for detecting base incorporation (to build a single channel sequencing-by-synthesis platform).
Virtual Wells
One of the key elements of the 454 and Ion torrent platforms was their use of wells to confine beads on an array (and in Ion torrents case over a sensor). Both these platforms used an off chip process to amplify DNA on the bead (emulsion PCR). This added an extra and somewhat awkward step to the sequencing process.
The Genapsys approach suggests doing away with wells completely. Instead magnetic and electric fields would be used to confine beads and reagents. Magnetic beads would be used which would be attracted to magnets on the chip, localising the bead over sensors.
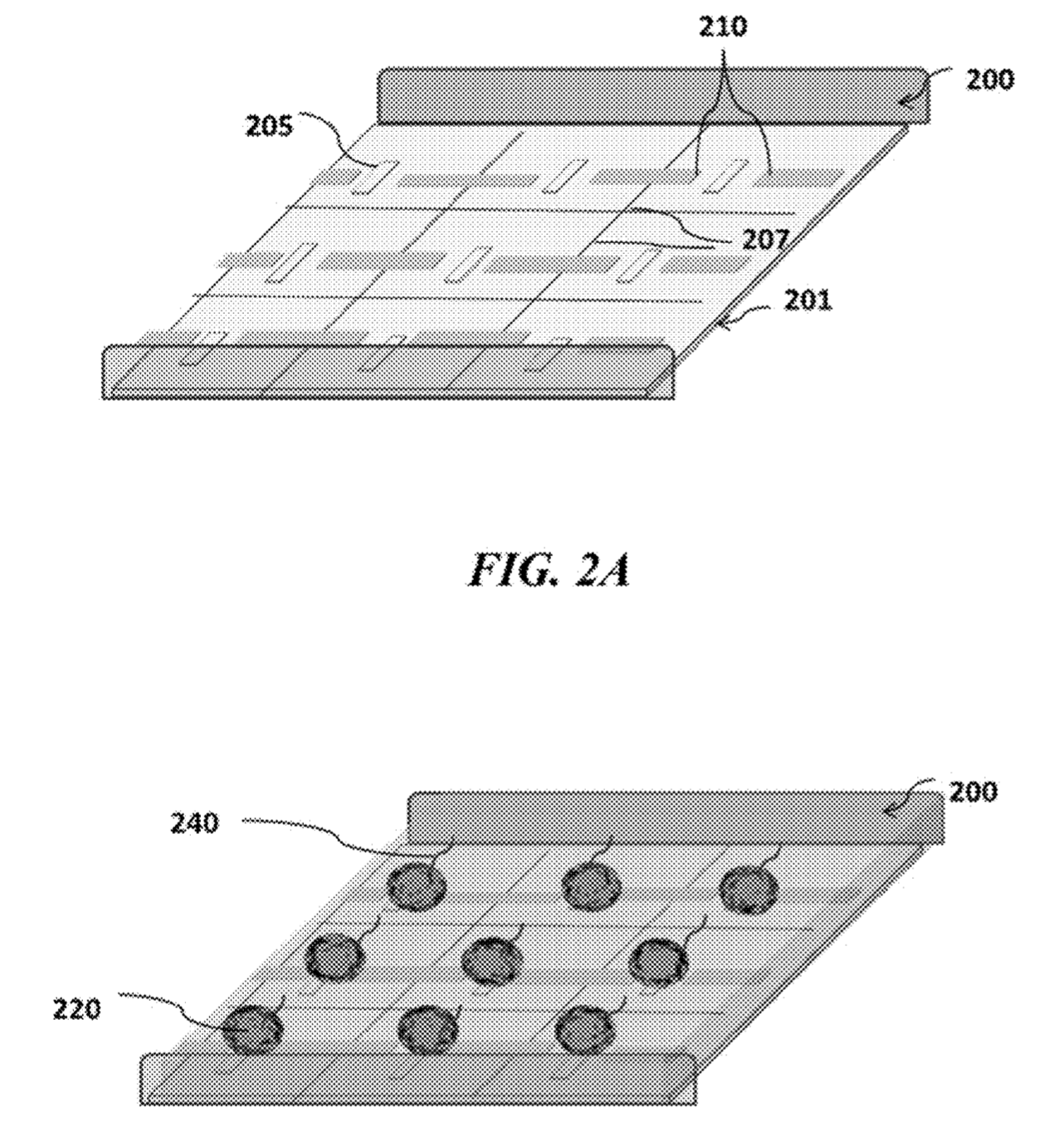
They then suggest using electric fields to confine nucleotides, strands, enzymes and potentially other reagents. Using the virtual well technology the amplification process could take place on chip. I assume this would be the same process as used for emulsion PCR. However, rather than the reaction vessel being a tiny water bubble, the charged reagents are confined by the electric field.
This seems like a rather neat idea, but I’m curious to know how well it works in practice. I would guess confinement is not perfect.
Detection
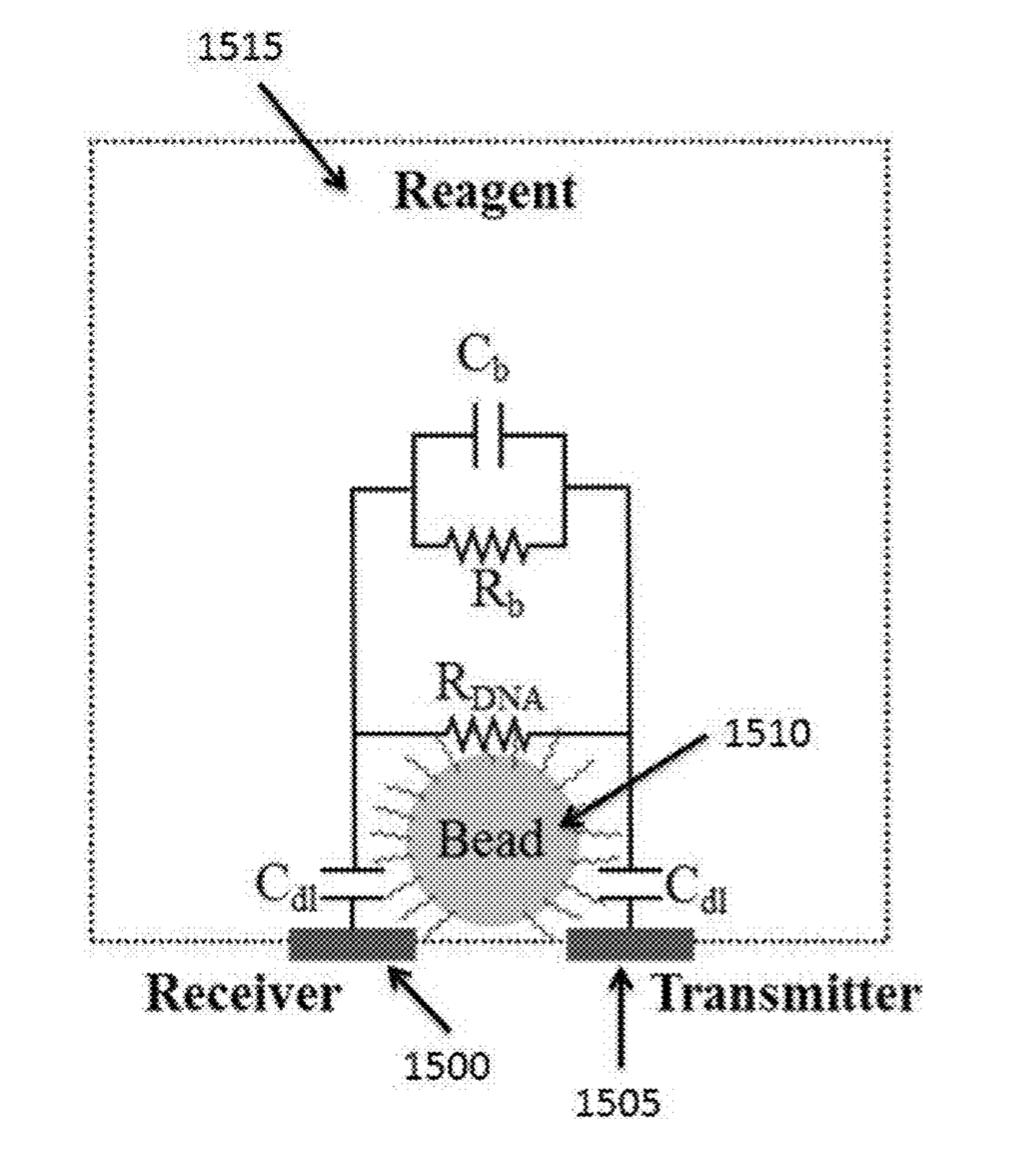
Initial reports all suggested that incorporation would be detected either through changes in pH (ISFET) or heat [2]. While recent patents still mention pH, heat, and charge based detection, they also discuss detection of incorporation through conductivity and impedance changes in the Debye layer of the bead [5].
The Debye layer as far as I can tell just means the double layer. My understanding is that in this scenario the bead will be negatively charged. There will therefore be a double layer formed around the bead:
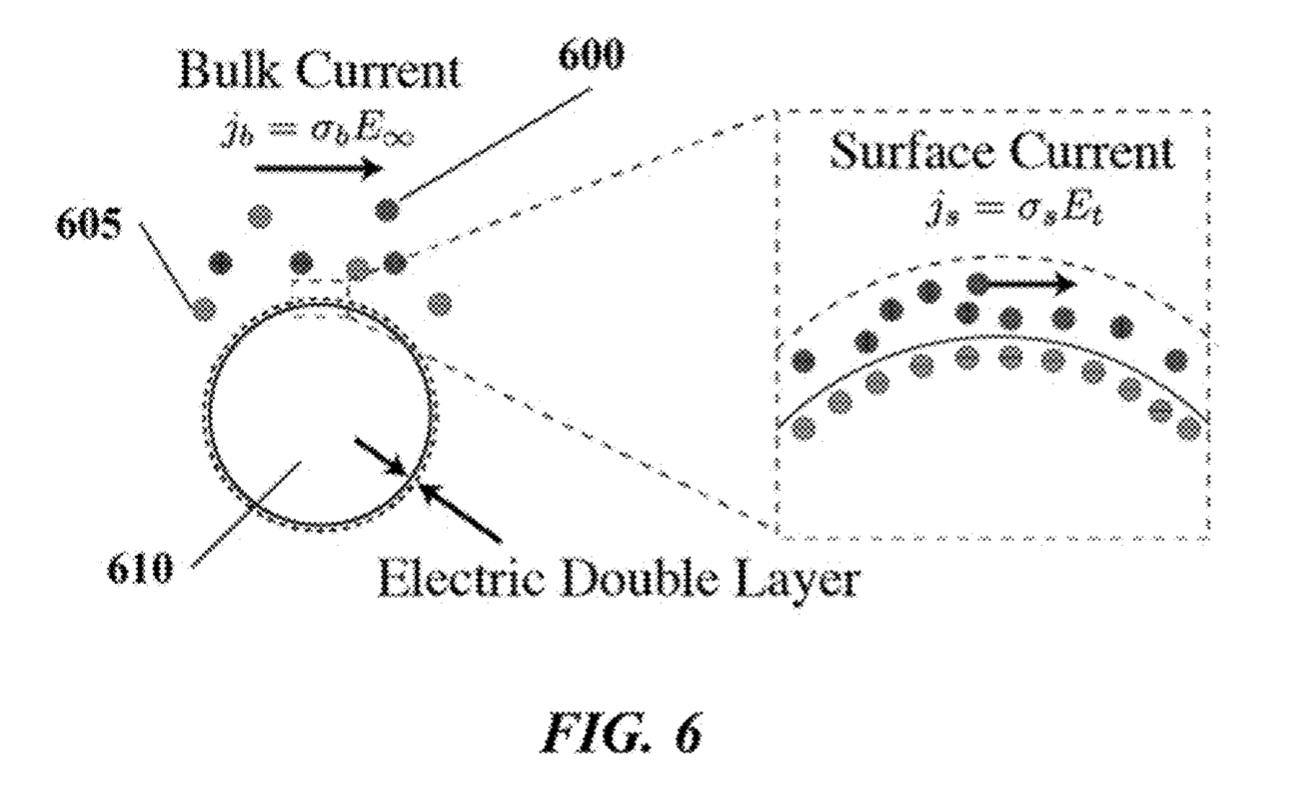
By placing electrodes on either side of the bead you can measure conductivity and impedance/capacitance. I would imagine that most current will come from the bulk of the solution, however the patents suggest that contribution from the double layer can be measured.
Exactly how additional nucleotides effect the conductivity/impedance of the double layer is less clear to me. As they will increase the negative charge in the vicinity of the bead it seems logical that they would however.
If it really is just the additional charge they add, the system feels similar to other methods of detecting the beads overall charge (like the caerus approach). However perhaps by looking at the change in double layer current background contributions are reduced, or the effect of the charge difference is amplified.
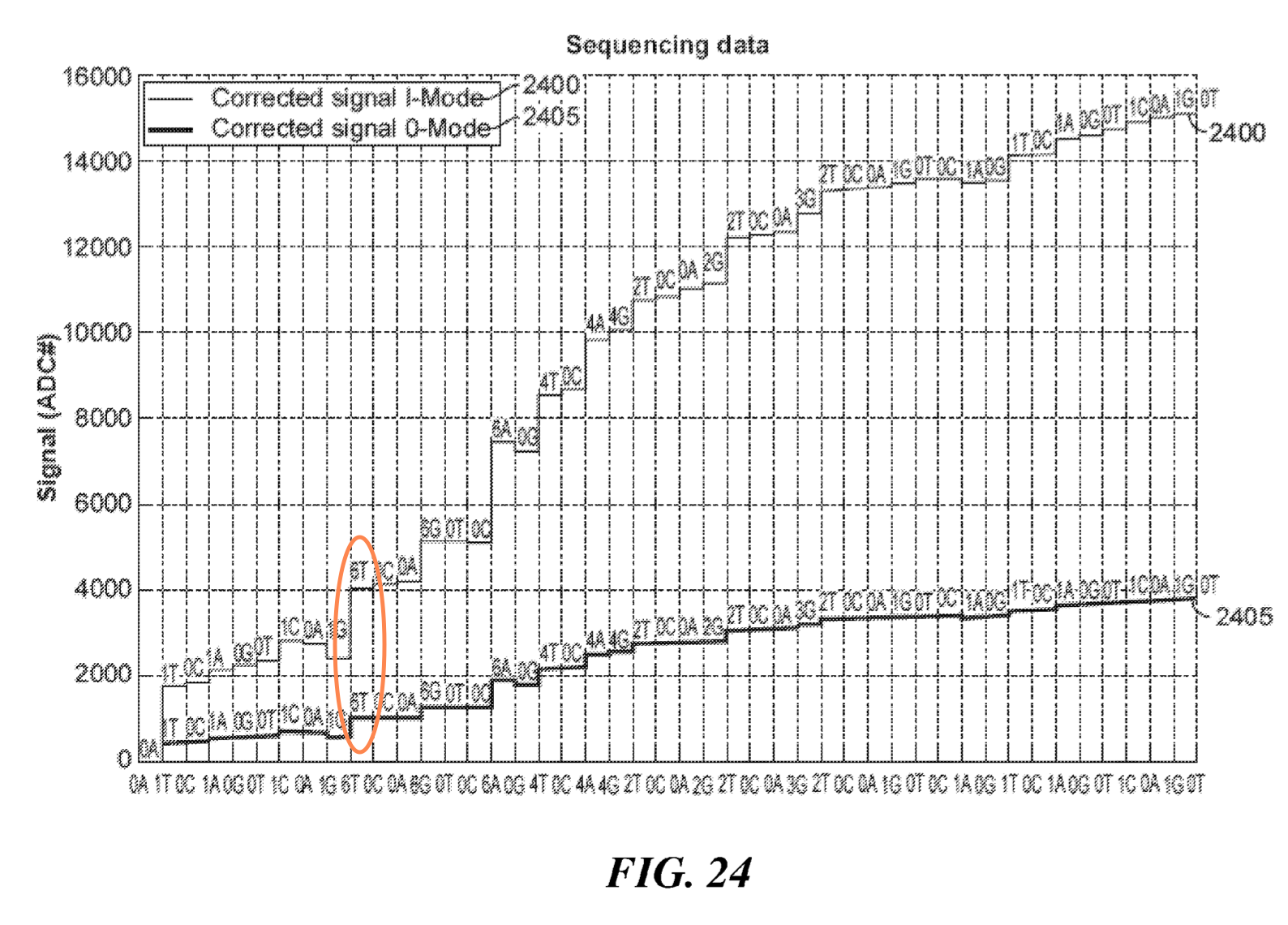
One patent does show “sequencing data”, as far as I can tell this is more likely to have come from simulation than a real experiment, I’ve highlighted a large deflection caused by multiple incorporation in the figure below:
Another factor that leads me to believe they may be pursuing a bead based, Debye layer based approach (rather than using ISFETs) is that pairs of electrodes are shown along side beads repeatedly.
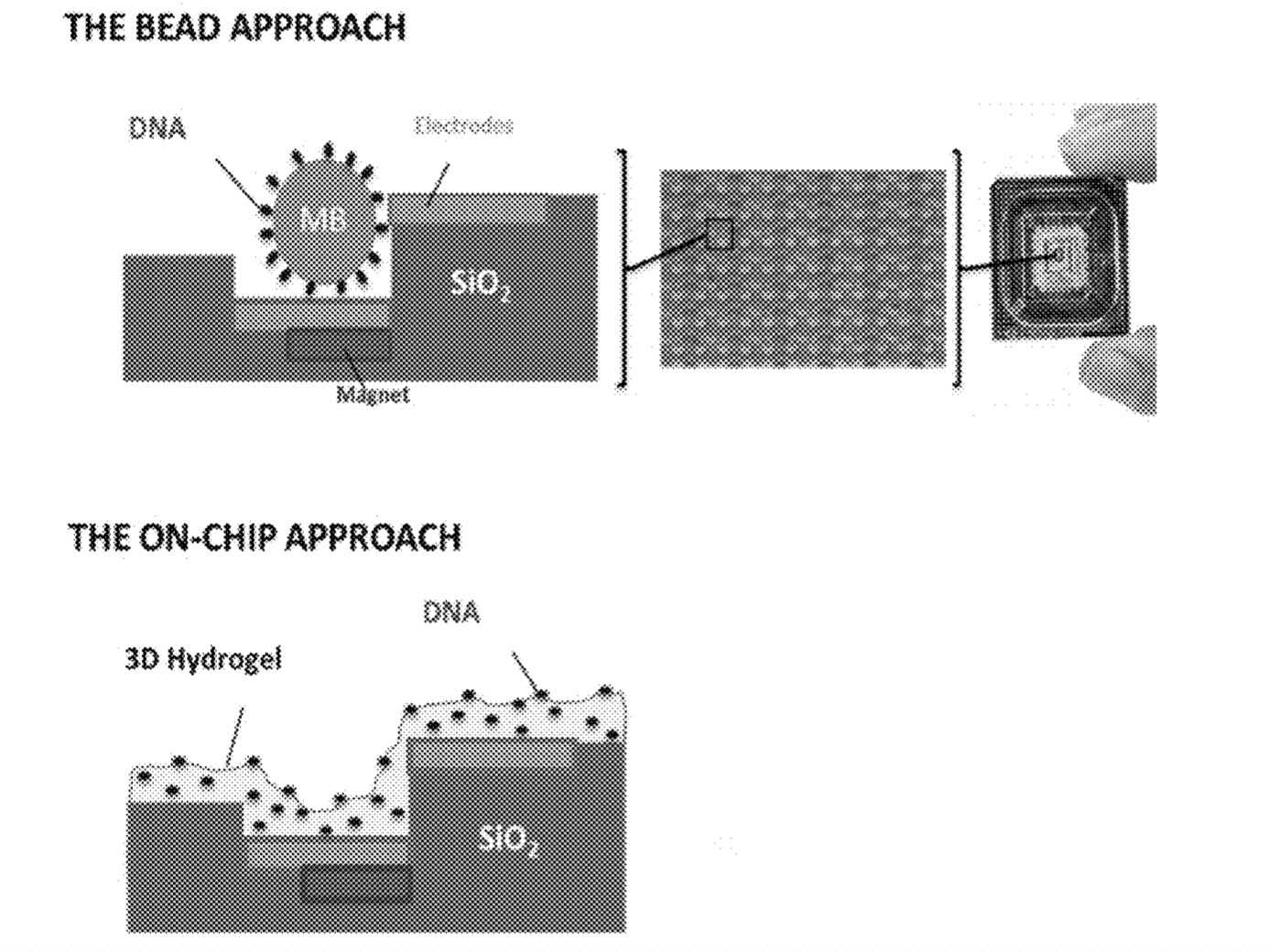
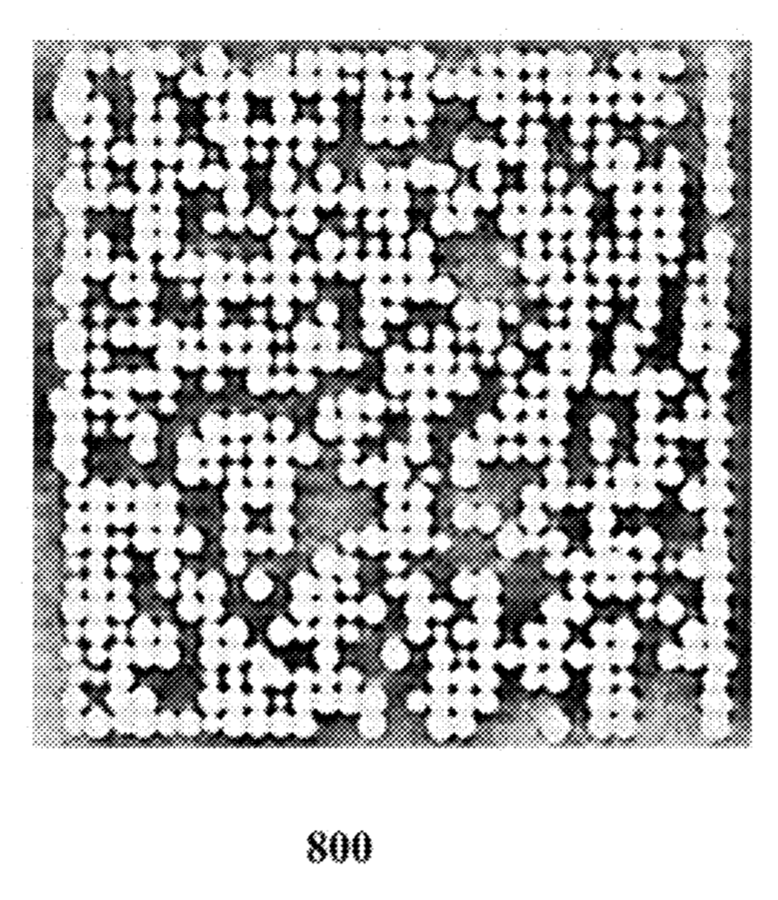
There are a few images of seemly real chip systems, but I didn’t see any SEM images. One example is this figure showing bead occupancy (which looks pretty good!):
 Overall, while Genapsys haven’t released a whole lot of public information, the patents seem to give a reasonable indication of what they’re working on. Charge based approaches seem attractive. One advantage they have over other chip based approaches is that you don’t need to monitor the incorporation in real time. Bases can be incorporated, and the charge difference measured at low speed, and potentially under different reagent conditions.
Overall, while Genapsys haven’t released a whole lot of public information, the patents seem to give a reasonable indication of what they’re working on. Charge based approaches seem attractive. One advantage they have over other chip based approaches is that you don’t need to monitor the incorporation in real time. Bases can be incorporated, and the charge difference measured at low speed, and potentially under different reagent conditions.
However, while patents suggest chips maybe reusable, you only get a single read from each “virtual well”. As with Ion torrent and 454, this could ultimately limit throughput.
Overall, some interesting ideas. And I look forward to seeing how things pan out.
Notes
[1] http://grantome.com/grant/NIH/R01-HG006889-03
[2] Genomeweb article: https://www.genomeweb.com/sequencing/genapsys-develop-microelectronic-sequencer
“DNA sequencing method that is based on direct heat or pH measurement”
US Patent No. 7,932,034, “Heat and pH measurement for sequencing of DNA.”
“In 2010, the firm won a $250,000 grant through the Qualifying Therapeutic Discovery Project Program for a project entitled “Development of an inexpensive, ultra-high throughput micro-electronic medical sequencer””
[3]
http://www.freepatentsonline.com/9399217.html
“As used herein, “virtual wells” refers to local electric field or local magnetic field confinement zones where the species or set of species of interest, typically DNA or beads, generally does not migrate into neighboring “virtual wells” during a period of time necessary for a desired reaction or interaction.”
Virtual Well
The a virtual well or “chamber-free array”, may detect or manipulate particles (e.g., beads, cells, DNA, RNA, proteins, ligands, biomolecules, other particulate moieties, or combinations thereof) in an array wherein said array captures, holds, confines, isolates or moves the particles through an electrical, magnetic or electromagnetic force and may be used for a reaction and or detection of the particles and or a reaction involving said particles. Said “virtual well” may provide a powerful tool for capturing/holding/manipulating of beads, cells, other biomolecules, or their carriers and may subsequently concentrate, confine, or isolate moieties in different pixels or regions of the array from other pixels or regions in said array utilizing electrical, magnetic, or electromagnetic force(s). In one embodiment the array is in a fluidic environment. Sensing may be done by measurement of charge, pH, current, voltage, heat, optical or other methods.
[4] 2013 patent: http://www.freepatentsonline.com/20130096013.pdf
ISFET, chemFET or nanobridge.
[5] Recent patent: http://www.freepatentsonline.com/y2018/0119215.html
“detect a change in conductivity within a Debye layer” ” can detect nucleotide incorporation events by measuring local impedance changes of the magnetic beads 220 and/or the amplified DNA (or other nucleic acid) 255 associated with the magnetic beads 220. Such measurement can be made, for example, by directly measuring local impedance change or measuring a signal that is indicative of local impedance change. In some cases, detection of impedance occurs within the Debye length (e.g., Debye layer) of the magnetic beads 220 and/or the amplified DNA 245 associated with the magnetic beads 220. Nucleotide incorporation events may also be measured by directly measuring a local charge change or local conductivity change or a signal that is indicative of one or more of these as described elsewhere herein. Detection of charge change or conductivity change can occur within the Debye length (e.g., Debye layer) of the magnetic beads 220 and/or amplified DNA 245 associated with the magnetic beads 220.”
“using the sensor to detect a change in conductivity within a Debye layer of the bead upon incorporation of at least one nucleotide of the population of nucleotides into a growing nucleic acid strand, which growing nucleic acid strand is derived from the primer and is complementary to the nucleic acid template”