Sony CCD-V30 Camera Viewfinder CRT Notes
Essentially the CRT module from this camera is very similar to that described in my previous post. Coils can be driven with +/- ~10V.
The image shows how power should be connected (5V).
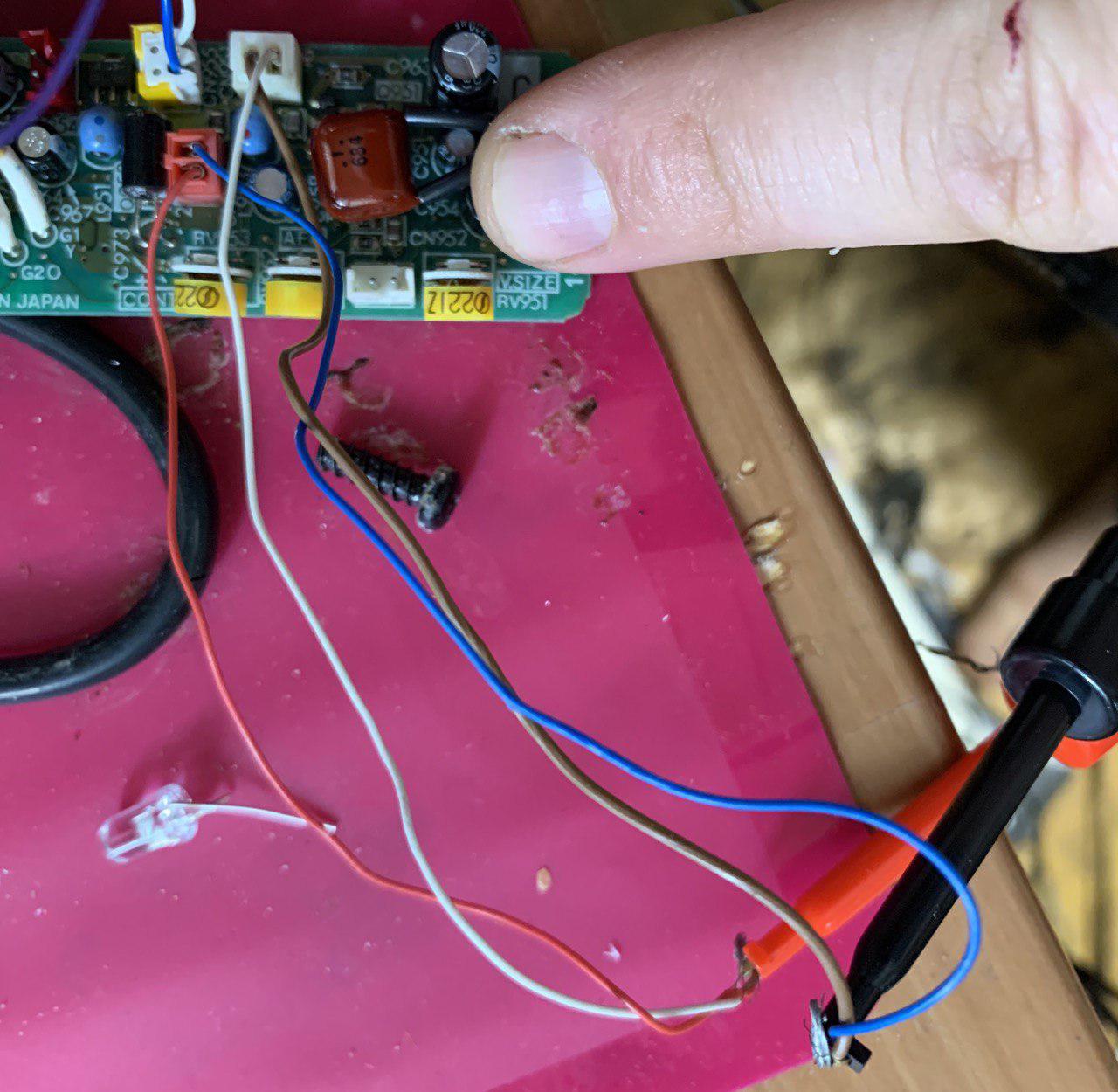

My contribution to the increasing entropy, and eventual heat death of the universe
Archive for the ‘Uncategorized’ Category.
Essentially the CRT module from this camera is very similar to that described in my previous post. Coils can be driven with +/- ~10V.
The image shows how power should be connected (5V).



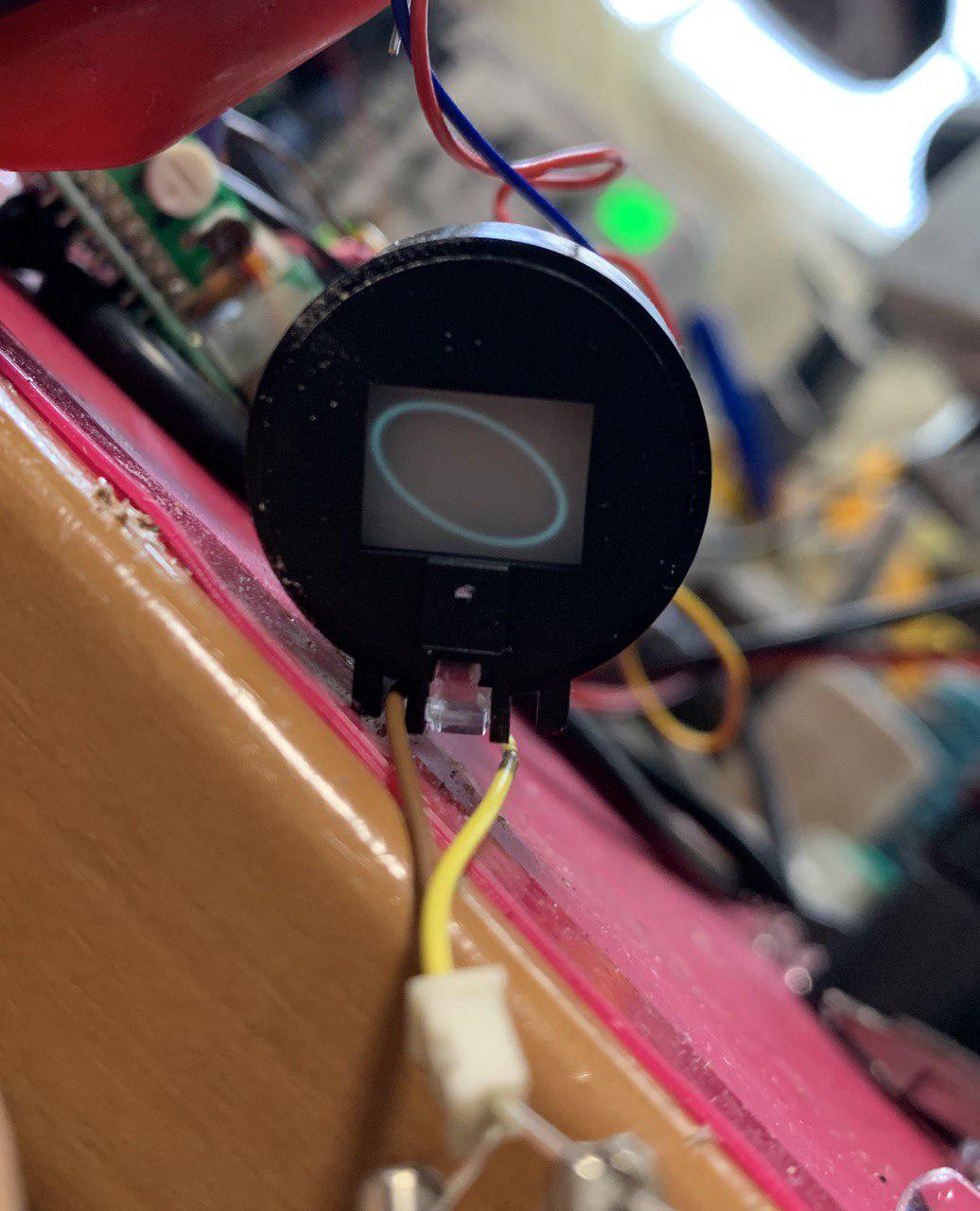
I was tearing down a Handycam and came across the viewfinder CRT. I have another project were I want to drive a CRT with an XY input and figured it would be good to experiment with this CRT module. These are my notes.
Here’s the module extracted from the camera:
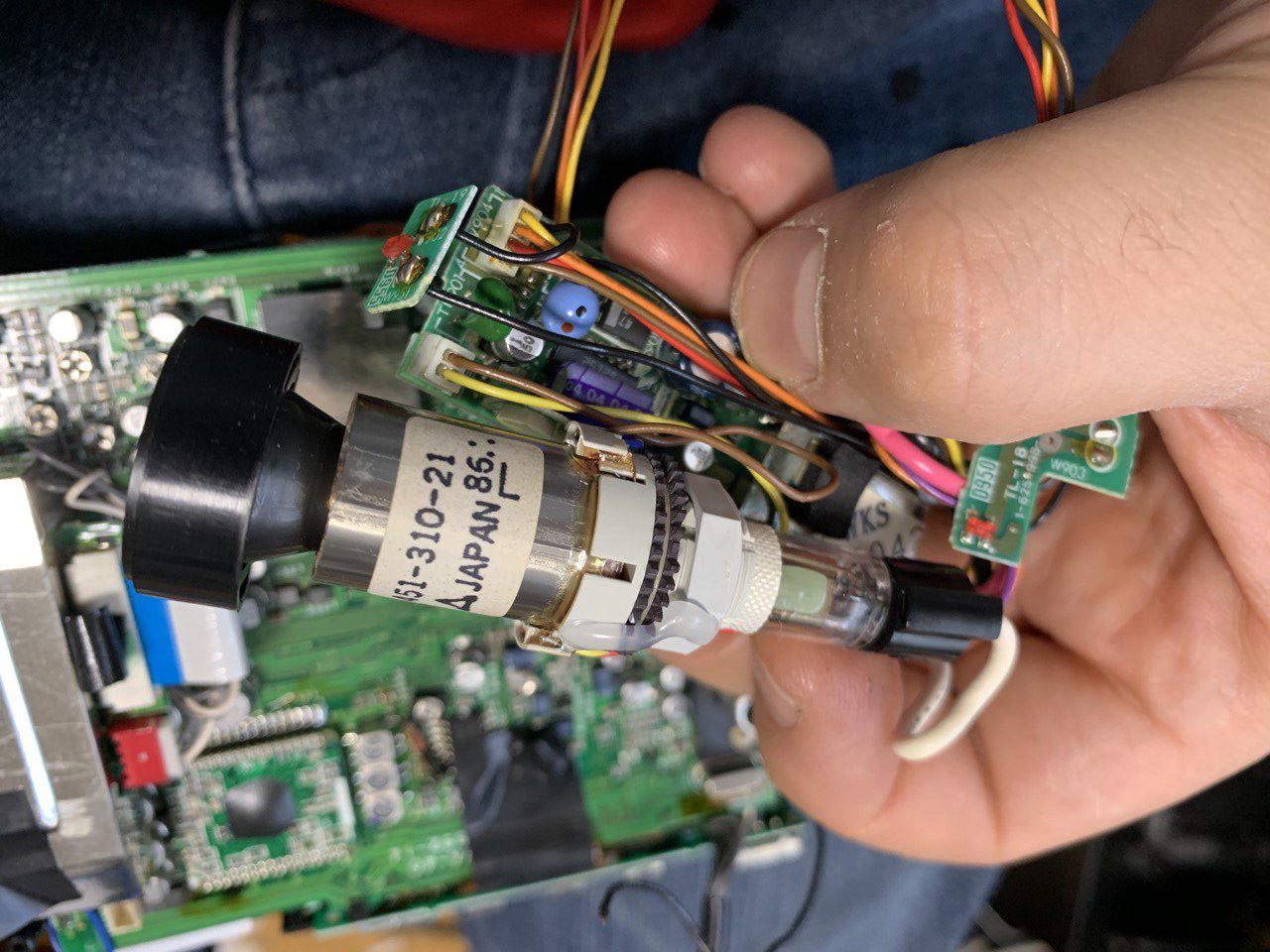
The module has a 4pin cable which connects it to the camera body. The module requires 5V power at ~60mA.
I fiddled around a bit to determine where power needed to be connected. The connector below the camera side. Clearly at least one of these pins should be signal input (my guess is the yellow wire). But the signal appears to need to be high/low to power up the module.

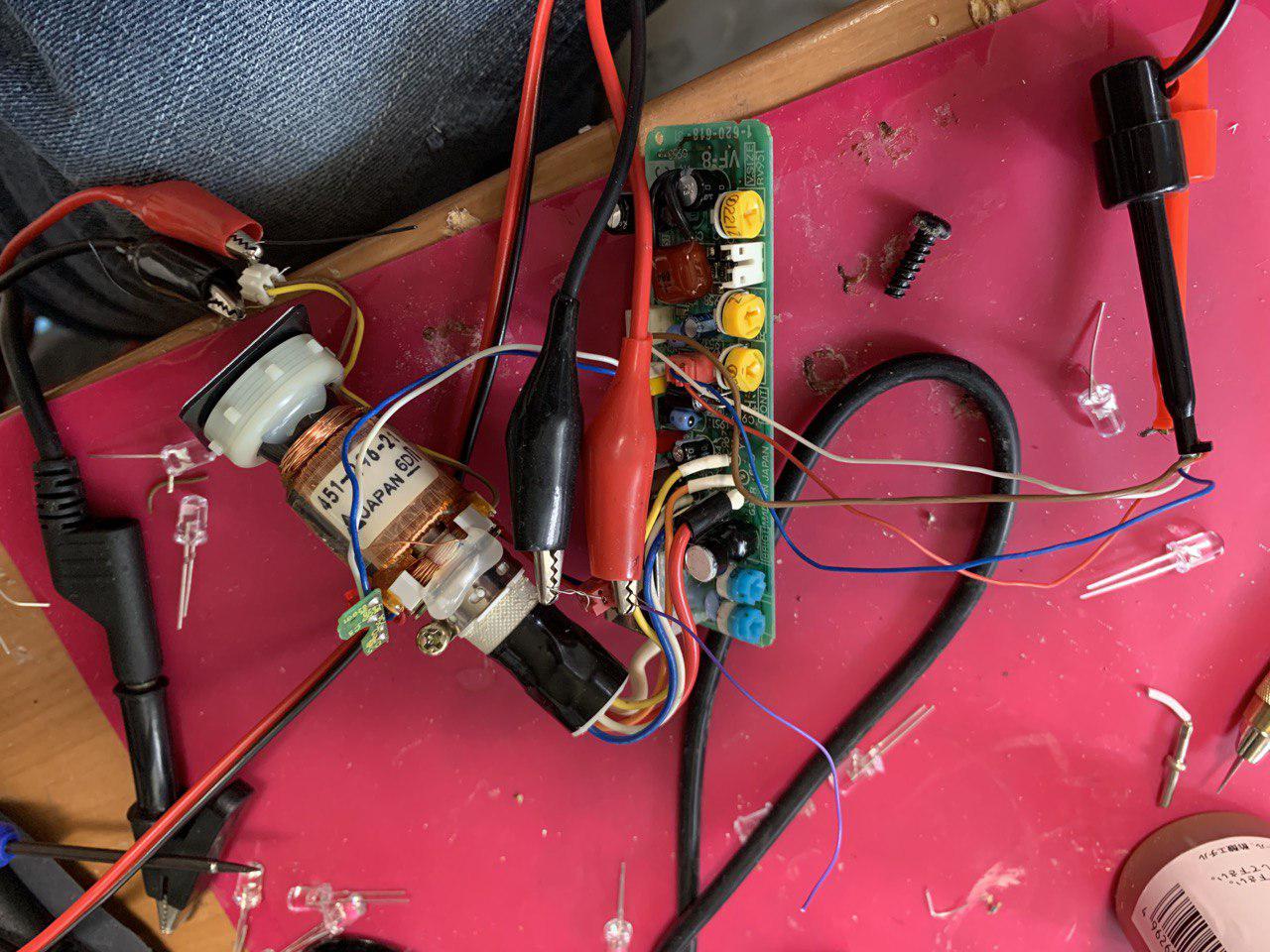
Below is the setup I used to drive the coils. I drive the coils directly from a function generator. 12Khz +/- 10V works well. The coils are essentially isolated from the rest of the circuit (as I understand it).

Fiddling with the pots on the module can help tighten the spot size, reduce the intensity. Below is a typical video. Couple of 12KHz +/- ~10V sine waves.
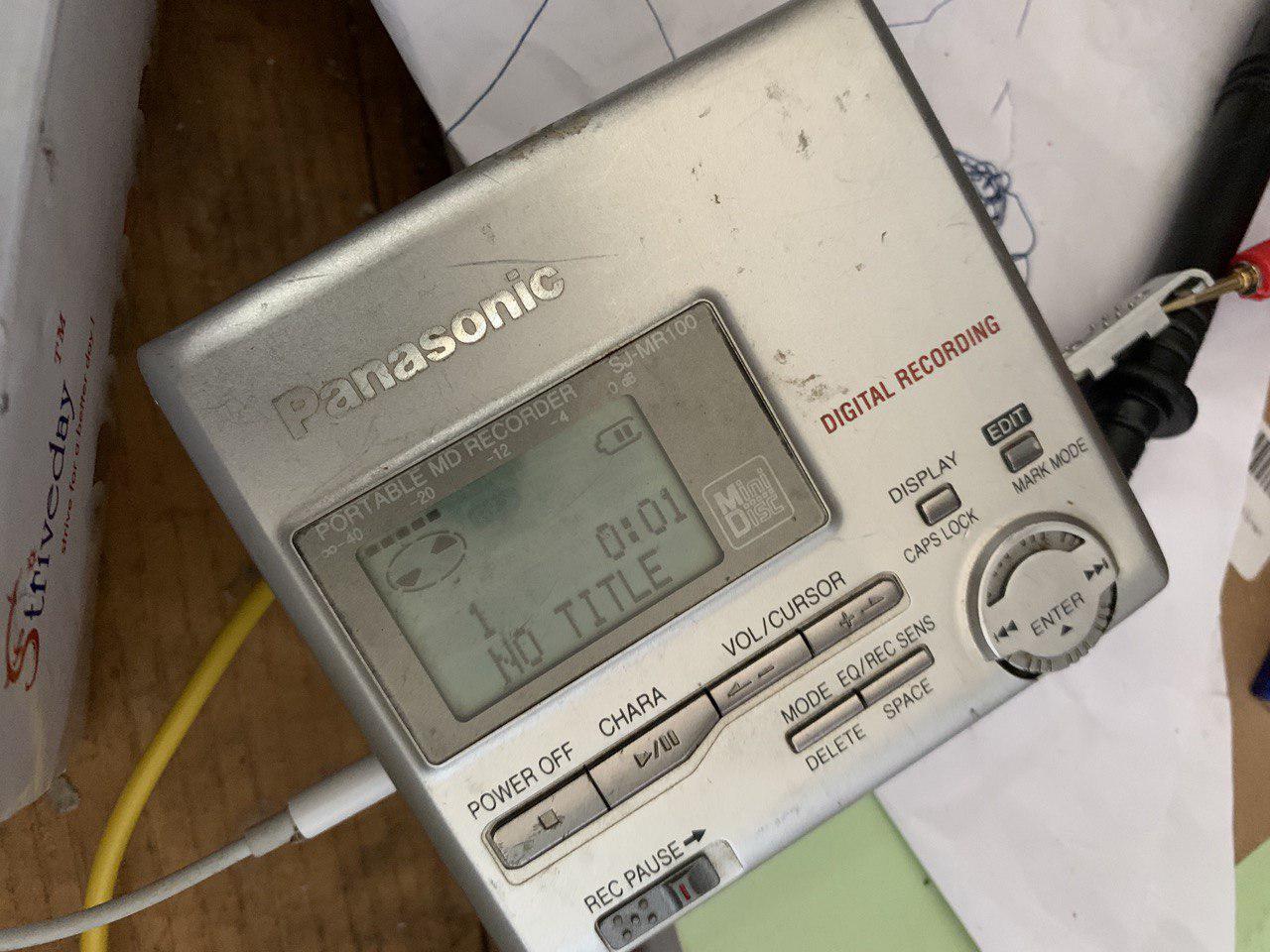
I picked up a Panasonic SJ-MR100 for 100 Yen in Akihabara about a year ago. It was sold as junk and came with no accessories. I figured it might be fun to play around with, and I have some minidiscs I recorded a few years ago I wanted to try.
The player/recorder came with no battery/charger. So I hooked it up to a bench supply. I used the internal “gumstick” battery terminals. You can hook the positive supply to the door and ground the internal part of the case. Not ideal, but works for testing. I used about 1.4V. 1.2V will trigger the low battery warning. It draws quite a bit of current when spinning up the motor, so a current limit of 700 or 800mA seems to be required.
When I tried my first few minidiscs they appeared to be playing, but there was no sound. At first I thought the drive was faulty. However, I now suspect this is because they were recorded as MDLP (I don’t know for sure, but I vaguely remember that I used MDLP frequently). The SJ-MR100 was released prior to the introduction of MDLP. It seems to try and play the disc but no audio is produced. The display shows the following (LP, mono):

During normal operation the display is as follows:
There’s a comprehensive review of this device which was hosted on aol. It’s unfortunately now only available via the waybackmachine. A pdf dump is here: http://41j.com/blog/wp-content/uploads/2020/05/Panasonic-SJ-MR100-Minidisc-Review.pdf
I also found the service manual online, local copy here:
One of the issues with testing for COVID19 and similar viral outbreaks is that kits can not be deployed ahead of time. Early in the outbreak the sequence of virus will not be known. Once determined a test specific to this outbreak will need to be designed, and then deployed. This is a significant logistical challenge.
Specifically, one of the more popular and accurate testing approaches to testing for COVID19 is real time PCR. The testing protocol is relatively straightforward. However, before kits can be deployed primers specific to the virus need to be designed and tested. They then need to synthesized and deployed. QCing and shipping potentially millions of oligos is a significant logistical challenge.
A sequencing based approach would avoid these issues. Viral extraction and sample prep kits could be deployed ahead of time. A complete sample could be sequenced, and aligned to detect the presence of a virus. We’d no doubt have contamination/background viral genomes. However, given the huge number of reads generated by current platforms this shouldn’t be an issue. Sequencing however is expensive compared with real time PCR. The cheapest complete runs are on the order of 500USD. It’s likely that we can multiplex samples. But this brings potential contamination issues, additional complexity ,the requirement that testing is centralized, and assumes that we can easily batch samples.
Based on the CDC recommended kits, Real time PCR of COVID19 appears to costs about 10 to 20USD in reagents per sample (as supplied by vendors).
Real time PCR is cheaper. Sequencing is more versatile.
Can we create a platform that is as simple and cheap as real time PCR, but doesn’t require primers specific to our viral sequence. One option would be to build a platform that synthesized primers on device. In such a platform kits could be deployed ahead of time. When a viral outbreak occurs primers would be designed and then then the sequence would be transmitted to instruments (over the Internet). Instruments could then synthesize the primers, and runs tests.
This would require a new approach to synthesis that could be integrated into a small diagnostic device.
However, another approach might be to combine aspects of sequencing-by-synthesis with real time PCR to selectively amplify regions in the target genome. In this approach we’d first want to cut the viral genome. We could use a restriction or nicking enzyme to cut the genome into smaller fragments at specific sites [1].
A procedure like Nick Translation can then be used to selectively amplify sequence around the Nick sites [5]. Normally in PCR we introduce all four bases at once. But in this approach we introduce labelled bases [3] one at a time, in the order that they appear in the target viral genome [2].
If the sample contains the target viral sequence, it will be fully extended by the nucleotides as presented in the correct order to extend this sequence.
If the sample does not contain the target viral sequence, it will be partially but not fully extended.
There will be a difference in fluorescence [3] intensity between the fully extended target viral sequences, and the non-target partially extended sequence.
As in real time PCR, the process would likely occur cyclically though rounds of melting while the fluorescence intensity [3] is monitored to detect amplification.
The above approach would result in a programmable detection system which does not require primers. Essentially the instrument would be a modified real time PCR instrument that could deliver nucleotides to the sample (in a programmable order) [4].
Notes
[1] I guess if there’s enough material (or it’s pre-amplified) we could also randomly fragment stick a polyA on the end and put our primers here.
[2] Reversible or other terminators, or another approach could be used here to ensure only a single base is incorporated into homopolymer runs. This approach is similar to that previously described to selectively amplify sequences here: http://41j.com/blog/2018/09/using-an-sbs-like-approach-to-selectively-amplify/
[3] Other detection methods than fluorescence could be used. We might use labeled, or unlabeled bases (perhaps ISFET detection of the incorporation of natural bases for example).
[4] We might need to attach the sample to a solid support for this to work. However, potentially we could put the sample in a sieving buffer. This would confine the longer fragments of the viral genome, while allowing single nucleotides to enter the matrix (potentially driven via electrophoresis). This concept is described here in more detail: http://41j.com/blog/2018/09/read-write-dna-devices-pt2/
[5] Or random sites, or sites as selected using a restriction enzyme etc.
….
CDC COVID19 Protocol: https://www.fda.gov/media/134922/download
2USD per sample: https://www.thermofisher.com/order/catalog/product/A15300
5USD per sample: https://www.qiagen.com/us/products/diagnostics-and-clinical-research/clinical-sample-processing/qiaamp-dsp-viral-rna-mini-kit/#orderinginformation
5USD per sample: https://www.qiagen.com/us/products/diagnostics-and-clinical-research/solutions-for-laboratory-developed-tests/ez1-dsp-virus-kit-na/?clear=true#orderinginformation
6USD per sample: https://www.qiagen.com/us/products/diagnostics-and-clinical-research/clinical-sample-processing/qiaamp-dsp-viral-rna-mini-kit/#orderinginformation