Random Notes 5
Xaar 128
https://github.com/MatthiasWM/Xaar128
https://github.com/gkyle/xaar128
https://www.xaar.com/en/products/xaar-printheads/xaar-128/
http://ytec3d.com/forum/viewtopic.php?t=47
chinese inkjet info: http://m.6lon.com/sdm/132165/2/cp-1003311/0.html

RNA Bioanalyzer
| Paper | Quantification pg/ul RNA | Run | Plots |
| Urine RNA Processing in a Clinical Setting: Comparison of Three Protocols | 397.5 ng/60 mL (Qubit RNA HS) | Agilent 4200 TapeStationRIN: 5.9 |  |
| Urine RNA Processing in a Clinical Setting: Comparison of Three Protocols | 397.5 ng/60 mL(Qubit RNA HS) | Agilent 4200 TapeStationRIN: 3.3 |  |
| Deep Sequencing of Urinary RNAs for Bladder Cancer Molecular Diagnostics | 0.98 ng/mL0.08 ng/mLProcessed within two hours of collection | Agilent 2100 Bioanalyzer, RNA Pico chipsRIN ranged from 2.5 to 9.5Mean: 6.03 | No plots |
| Identification of microRNAs in blood and urine as tumour markers for the detection of urinary bladder cancer | ~2 ng/uL (fig. 1) | Several urine RNA isolates showed low RIN values <3 | No plots |
| Urinary MicroRNA-Based Diagnostic Model for Central Nervous System Tumors Using Nanowire Scaffolds | Qubit microRNA Assay | Agilent 2100 BioanalyzerRIN: 8 |  |
RNA Papers
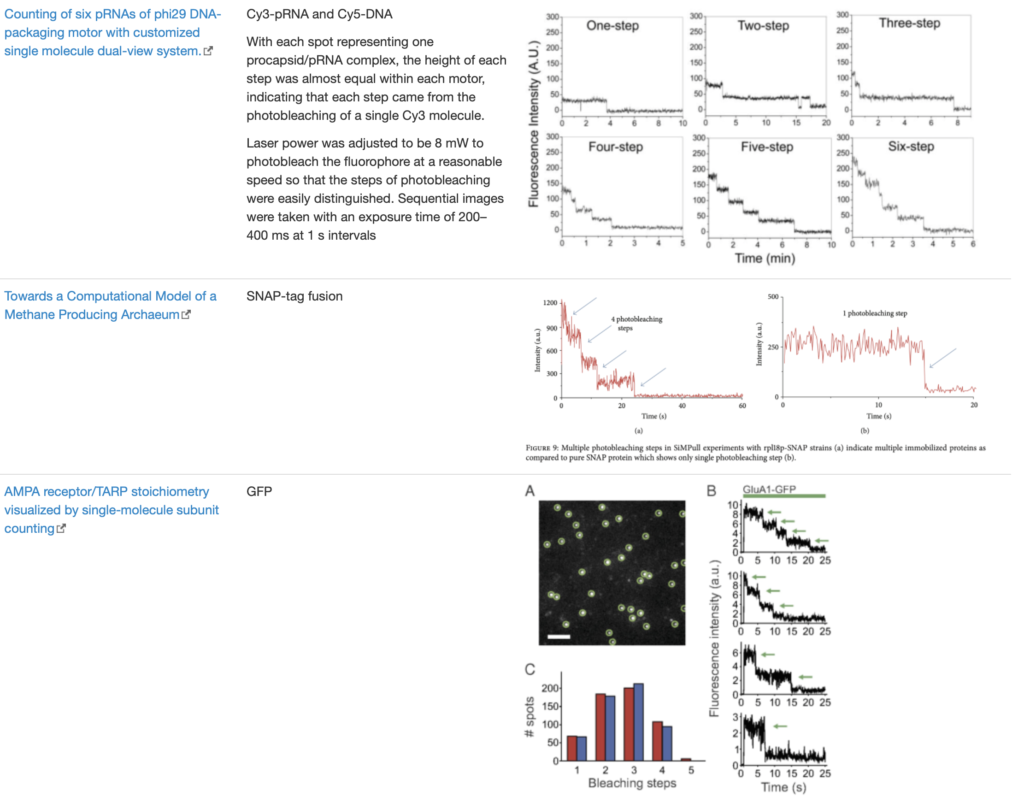
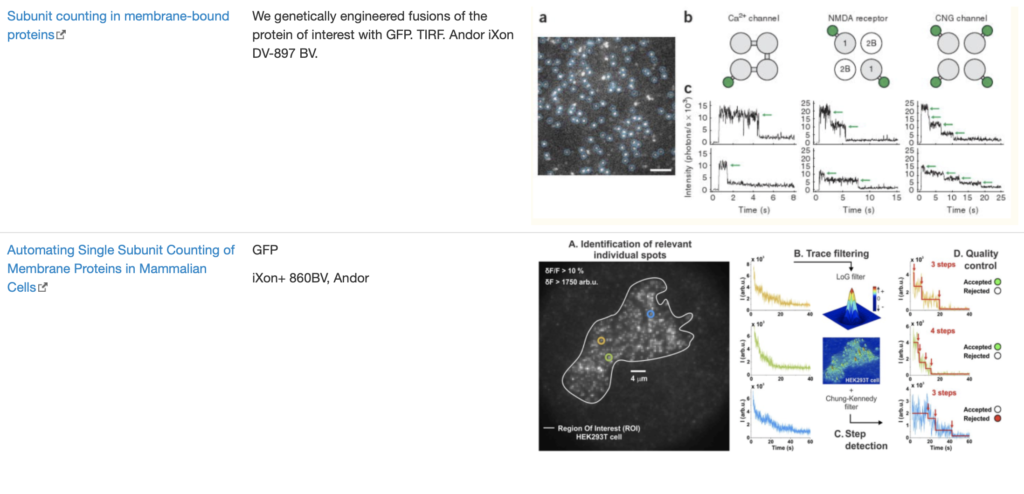
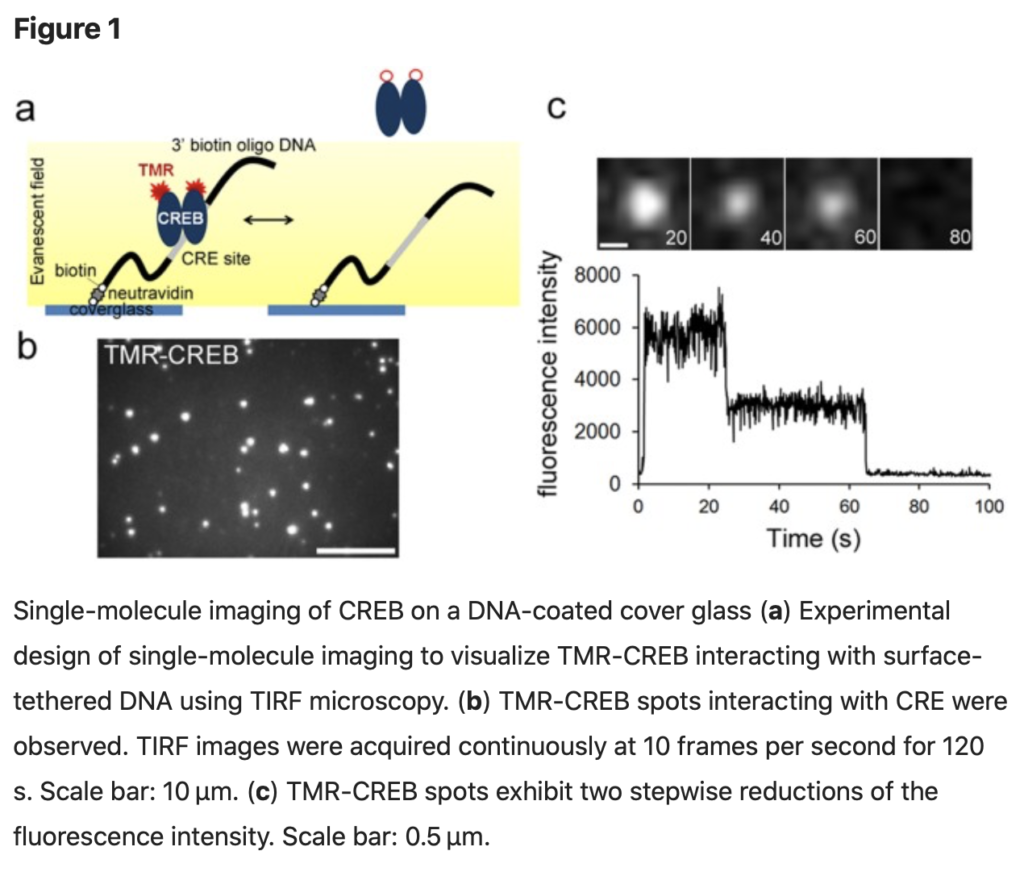
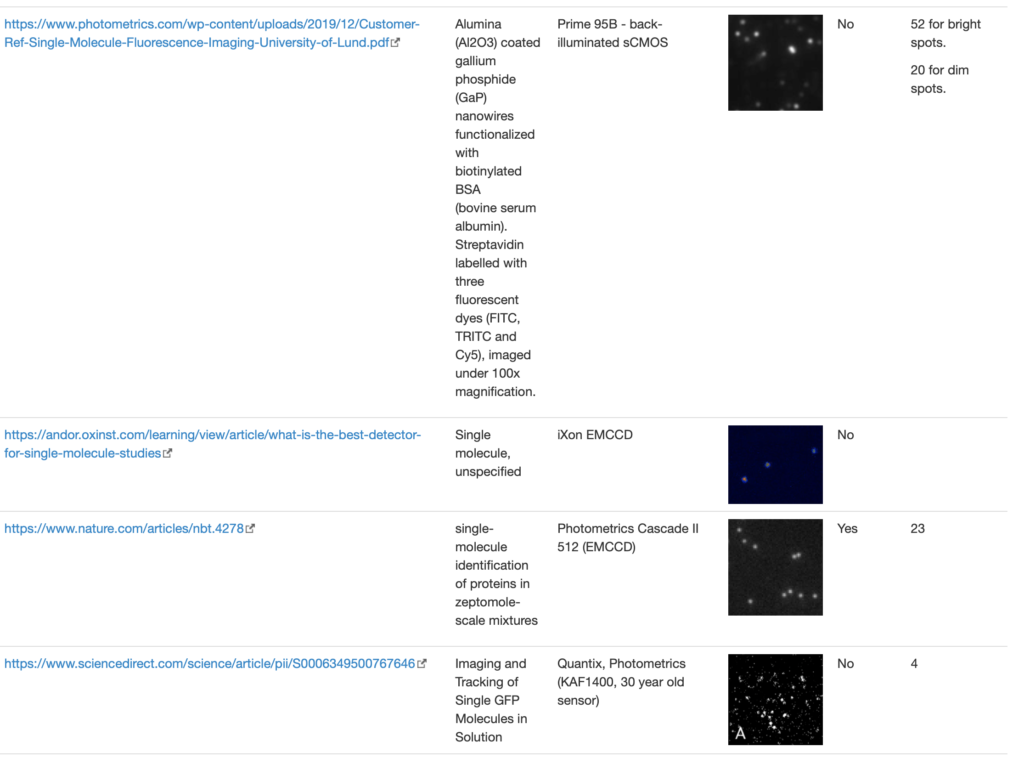
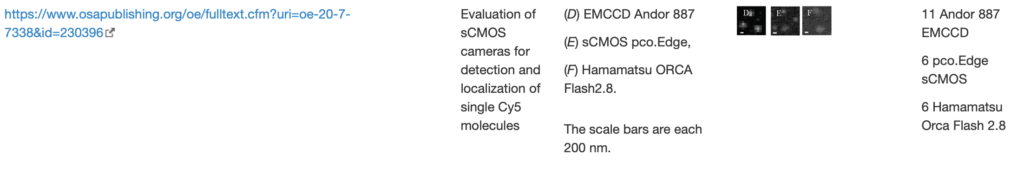
SingleMol Imaging
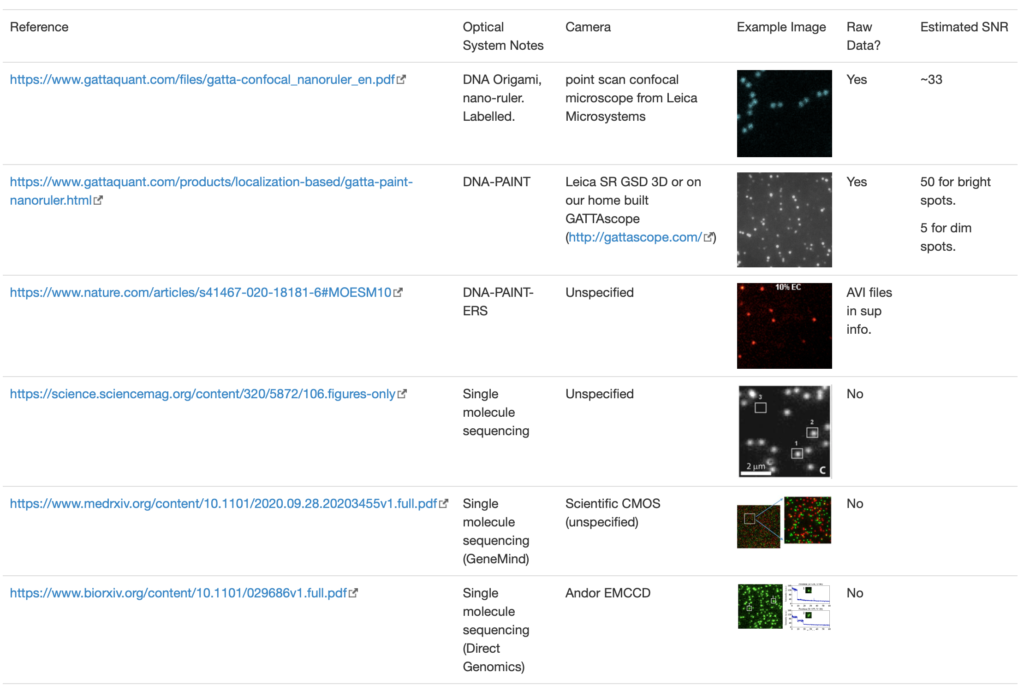
Solid state Nanopore
Hitachi, making pores with dialectric breakdown, slowing, notes on viscosity to slow etc: https://www.nature.com/articles/srep31324.pdf
Keio University group, optical observation of translocation: https://iopscience.iop.org/article/10.7567/APEX.9.017001/pdf
Amplification/Electronics
ASIC Design, to go to 1MHz: https://www.nature.com/articles/nmeth.1932
Noise measurements from above:
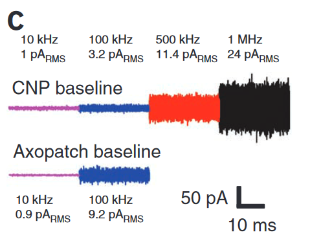
Instrumentation for Low-Noise High-Bandwidth Nanopore Recording. Engineered Nanopores for Bioanalytical Applications: https://www.sciencedirect.com/science/article/pii/B9781437734737000030
Above has lots of nice protocol information.
Ionic Speed/Current
“If we recall that an Ampere corresponds to a charge flow of 1 Coulomb each second and further, use the fact that the charge on a monovalent ion is approximately 1.6 x 10-19 Coulombs (that is 1 electron or 1 proton charge), then we see that a current of about one pA corresponds to roughly 107 ions passing through the channel each second. This value is in agreement with measurements” http://book.bionumbers.org/how-many-ions-pass-through-an-ion-channel-per-second/
Molecular Dynamics of Ionic Current: https://www.ks.uiuc.edu/Research/silica/IonRect.html

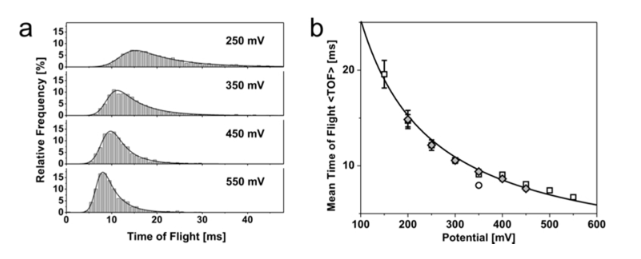
DNA Speed:
From, measures time to translocate two adjacent pores. distance is ~1.5um: https://pubs.acs.org/doi/full/10.1021/nl2030079
Folded/Knotted DNA:
Detection of knotted DNA
https://www.nature.com/articles/nnano.2016.153
https://www.nature.com/articles/s41467-019-12358-4
Folding
https://www.ncbi.nlm.nih.gov/pubmed/20608744
Double stranded Lambda DNA (16um long, 48kb). Translocates in 3 different confirmations. Grabbed at the end. Grabbed in the middle (completely folded), Grabbed near an edge (partly folded). Difference in current levels is ~1nS between each confirmation (200pA). 22nm pore is used. Translocation time ~3ms.
Hairpins, P2000 pulled quartz nano-pipette. Using hairpins to encode DNA 8bp and 16bp hairpins.
DNA Folds for data storage by Ulrich: https://www.ncbi.nlm.nih.gov/pubmed/30585490