Roswell Revisited
This post previously appeared on the substack.
I’ve previously talked about Roswell. But they’ve recently been making a bit more noise, and I thought It would be interesting to review my previous post in light of this new information.
Roswell was founded in 2014, back in 2018 they were reported to have raised $6M. Cruchbase now lists them as having raised $37M though it’s not clear how recent this figure is. A recent Genomeweb article says they now have 50 employees.
Roswell’s patents show a basic configuration where a strand of dsDNA is suspended between two electrodes as a kind of scaffold. A polymerase is then attached to this scaffold. The polymerase performs synthesis of a template strand and conformational (and other changes) are detected:
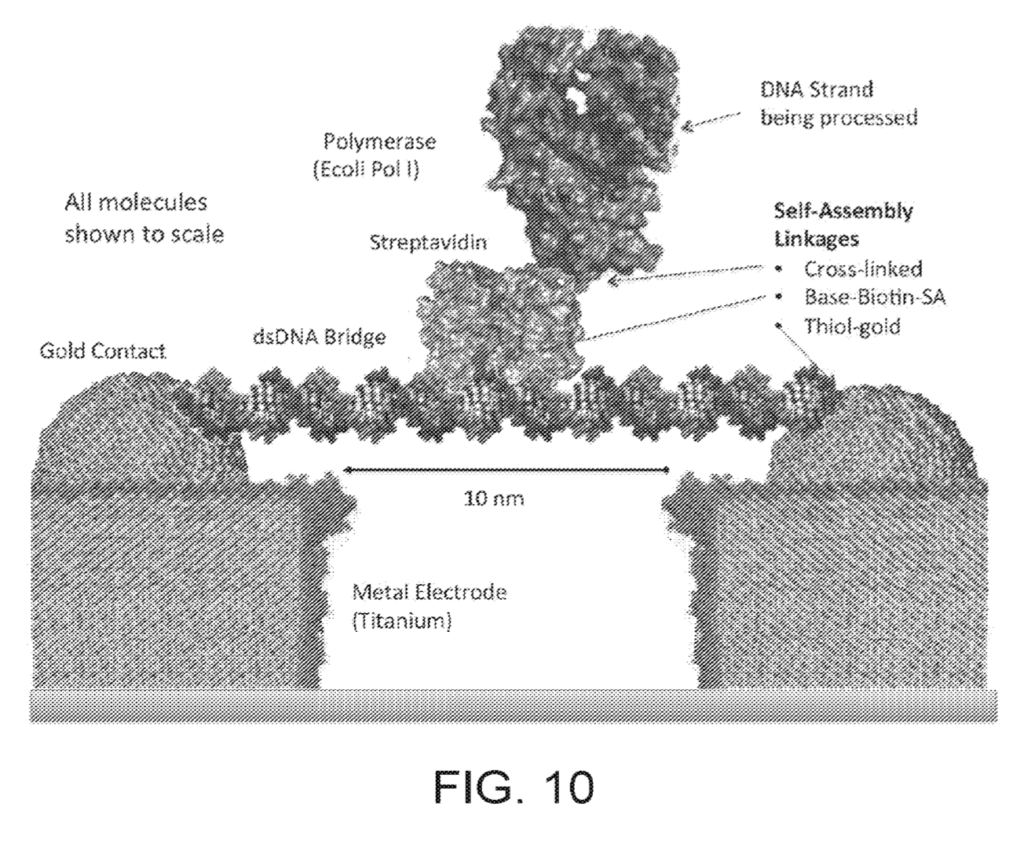
Detection is likely through tunneling (and potentially other) currents between the electrodes. Conformational changes in the polymerase and/or incorporated nucleotides would result in current changes that could be detect to sequence the strand under synthesis.
The patent I previously looked at showed data from homopolymers, with current defections on the order of 60pA.
So what’s new? Well, if the animations on their website are anything to go by, the basic mechanism is still broadly as described above:

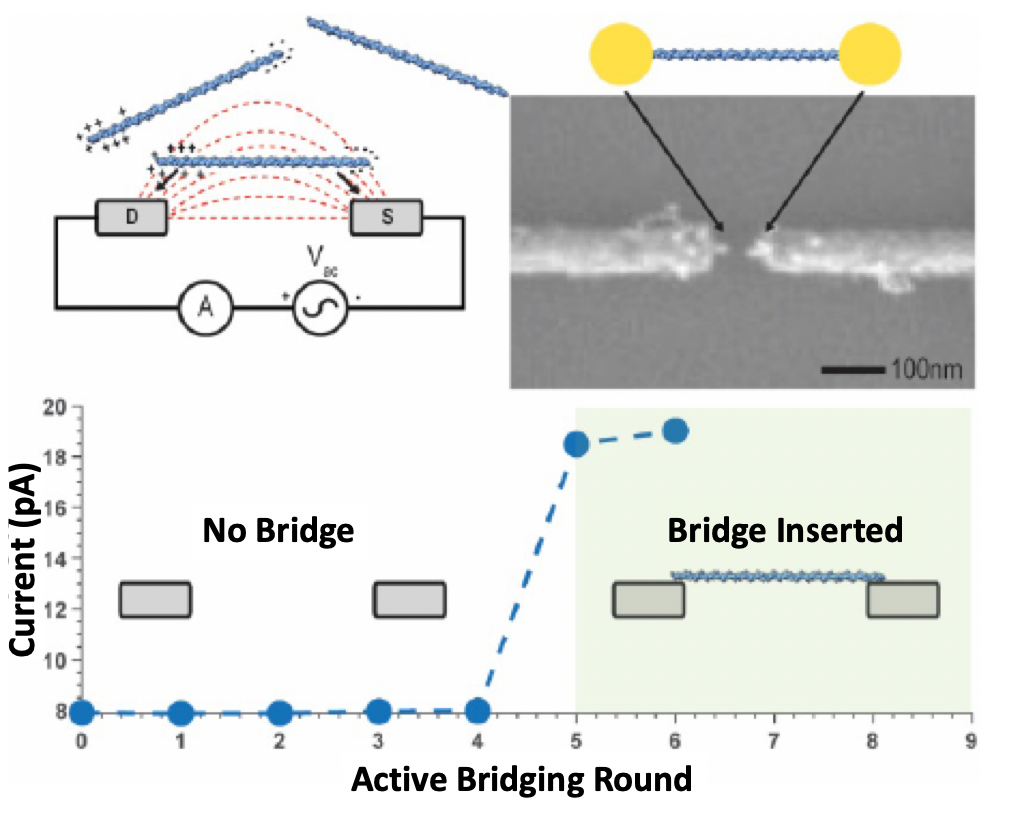
Roswell’s whitepapers also help clarify this process, where an electric field is used to attract the bridge molecule (dsDNA). The approach seems pretty neat, as you can turn on the field, attract and attach the dsDNA and then immediately turn off the field. This means that you should be able to attach a single bridge molecule to each sensor. Without something like this, you would be Poisson limited, with many sensors having no, or multiple bridges.
The Genomeweb article suggests they’re shifting away from DNA sequencing and “targeting diagnostics, especially for infectious diseases, or drug discovery as its first applications”. This is perhaps because their sequencing accuracy isn’t quite competitive yet:
“”Discrimination of the four bases can approach over 90 percent, and in complex templates, 70 percent,” Mola said.”
It’s difficult to unpack this statement without knowing exactly what experiments were performed and how the data was processed. But to me it suggests they’re not quite sequencing yet, but rather looking at ensemble properties of synthetic templates.
The white paper also tends to focus on applications other than DNA sequencing. For example hybridization based assays:
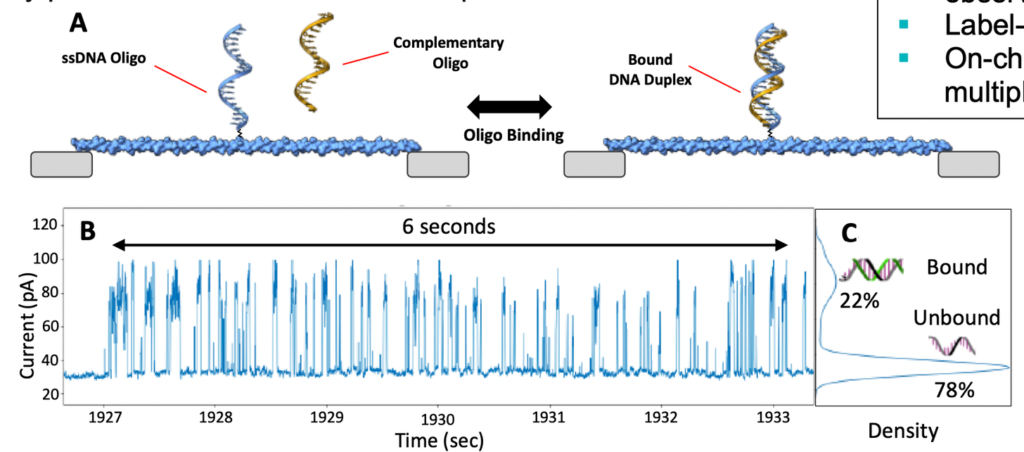
I personally find these other applications less compelling than sequencing. The advantage over other approaches, including other single molecule approaches, is unclear to me. But I do find the fact that the approach works at all, kind of exciting.
The other nice feature of the Roswell approach is that they can potentially pack features much closer than you can on a protein nanopore chip. The density they show, of 2500 sensors per mm² seems pretty good.
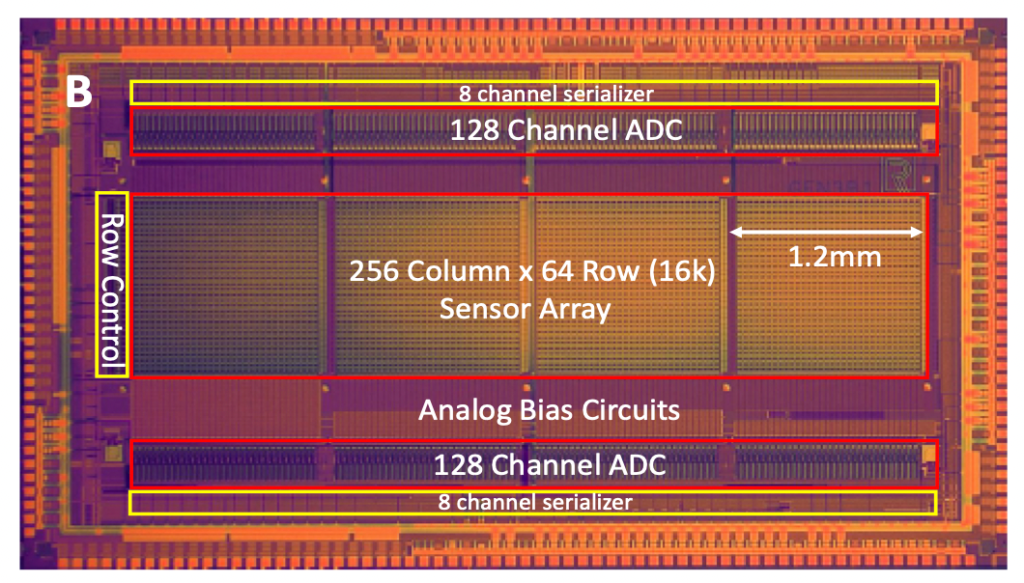
This is an advantage because it keeps the chips relatively small. Roswell’s Gen3 chips are 4x6mm. When I last looked at image sensor pricing, it seems like chips of this size would generally retail for about $10.
That’s pretty cheap, but still an expensive COGS for a consumable, which given the approach I suspect is not washable/reusable. This potentially makes the approach unsuitable for low-cost diagnostic applications.
However, that their approach works at all is neat, and I’ll be watching Roswell’s progress with interest.
Disclaimer: As always, you should be aware that I have equity in sequencing companies (based on prior employment) and am current working on a novel sequencing approach.