The Centrillion VirusHunter
DISCLAIMER: I’m currently working on a seed stage sequencing related project. So, keep that in mind when evaluating my comments. And get in touch ([email protected]) if this might be of interest.
There’s a new press release out from Centrillion suggesting their sequencing platform should be available now/soon. I’ve previously covered Centrillion, and recommend referring to that post for background on the company.
The press release doesn’t give much information, but references a recent publication, which provides more details. The paper describes a tiling array covering the entire SARS-CoV-2 genome. This is essentially a traditional microarray which covers the every possible SNP in the known SARS-CoV-2 genome. They describe this as follows:
“Here we describe a full genome tiling array with more than 240 000 features that provide 2x coverage of the entire SARS-CoV-2 genome and the use of such a genome tiling array to sequence the genome from eight clinical samples”
“Each base has two corresponding probe sets: one for the sense strand and one for the antisense strand.”…”one for each base”
So, Centrillion use a total of 8 probes per site. SARS-CoV-2 has an ~30Kb genome, which gives us the 240K features mentioned above. They use 25bp strands in their array. Tiling arrays have a number of limitations as compared to sequencing based variant detection. Firstly, it’s not clear what issues might be caused by multiple mutations covered by the 25mer. Such mutations would likely reduce hybridization efficiency, and could result in a variant miscall or “nocall”. The approach is also not able to detect deletions or insertions. These limitations make tiling arrays less interesting as a general purpose tool.
I personally would not characterize tiling arrays as “sequencing” or “resequencing”. In a tiling array you get a single read out for each position. Sequencing approaches generally produce a continuous readout of bases, without prior knowledge of the sequence context.
The paper presents a comparison of their approach against Illumina sequencing, summarized the in table below:
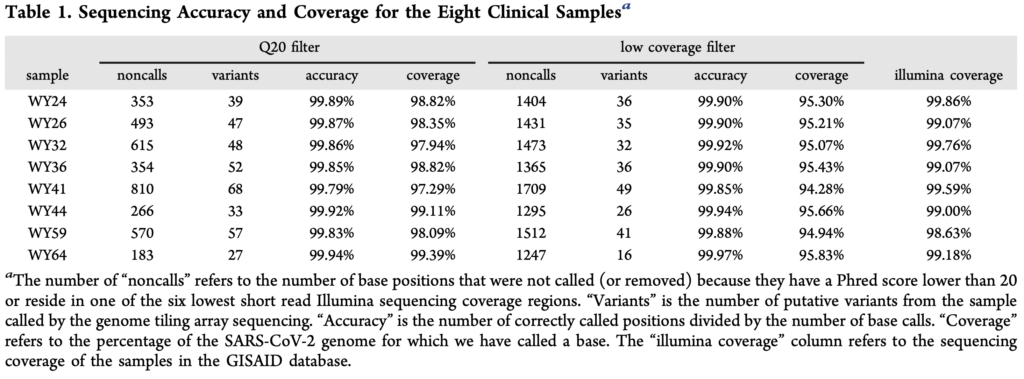
From the table above, noncalls are in the region of 1% seems to be one of the more concerning issues. I assume this means that both sets of probes essentially failed to provide a useful signal at these positions. The accuracy for the remaining positions seems reasonable (though less than I’d expect, given a known reference for the exact variant).
Summary
This is a fairly traditional tiling array for SARS-CoV-2 variant detection.
From the press release, it seems this is pitched as a lower cost alternative to sequencing. I don’t have good current pricing for microarrays, but I imagine the array they’re suggesting would cost >>$30, based on microarray prices I’ve seen. This is significantly more expensive than qPCR based testing (probably x10). This makes it too expensive for routine use in SARS-CoV-2 testing.
A detailed cost comparison against sequencing would be interesting. But I suspect we’re not talking about an order of magnitude cost difference. For me, this wouldn’t be enough to make an array based approach attractive, given that sequencing provides a richer dataset and more accurate.
Hopefully Centrillion will continue to develop some of their sequencing based ideas in the future.