Eve Biomedical
Today I’m going to briefly review a slightly less well known DNA sequencing company, Eve Biomedical. Checkout the complete list of sequencing companies, for links to all other posts.
Business
Eve Biomedical is a Californian company (C2934369) founded on 11/13/2006. They received ~1MUSD of SBIR grants, starting in 2013. Crunchbase lists them as having raised 7.7MUSD (from DFJ) most recently in December of 2012. I could find only 3 current employees on LinkedIn, and around 10 previous employees.
Technology
I could find 2 patents assigned to Eve Biomedical [1]. I’m going to focus on the earliest, because it’s the most fascinating to me [1a]. The patent describes a process they call “Rotation-dependent transcriptional sequencing”.
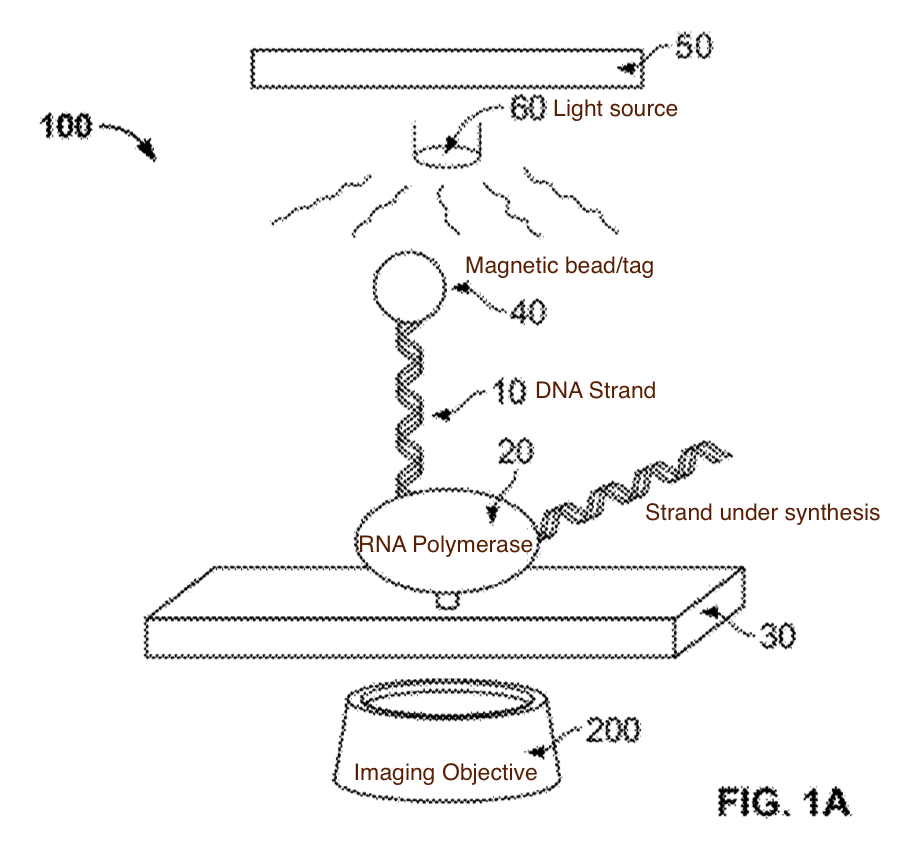
The sequencing approach relies of the fact that as an RNA polymerase incorporates a base, it rotates the template DNA strand. The approach is therefore sequencing-by-synthesis. However rather than synthesising a complementry strand of DNA (as in Illumina and other approaches) DNA is being transcribed to RNA and we are detecting the incorporation process as it takes place.
How to we detect the rotation of single strands of DNA? Stick a bead on it… An asymmetric magnetic bead is used. This allows us to put it under slight tension using a magnetic field (so it doesn’t move around under brownian motion). The patent shows 2.7 micron beads, it’s slightly surprisingly to me that a single polymerase/strand is able to move what I assume is a comparatively huge bead (if anyone has dimensions for polymerases I’d be very interested!).
A schematic of the system (from the patent) which I’ve annotated is shown below:
With this basic system in place, there are a couple of questions remaining. The first is, what exactly do these tags look like. The patent describes tags that look like a bigger bead with a smaller one stuck to it, I didn’t look into the construction details, but I’d imagine these can easily be fabricated:
The second question, is how to we use these parts to build a DNA sequencing platform.
If we are able to detect incorporation events, it’s hopefully clear that we can use this system to sequence DNA. One option is to simply flow bases in one at a time, and detect when bases are incorporated. This approach is similar to other single-channel, unterminated sequencing-by-synthesis approaches (Direct Genomics comes to mind, Ion torrent would be a related bulk approach).
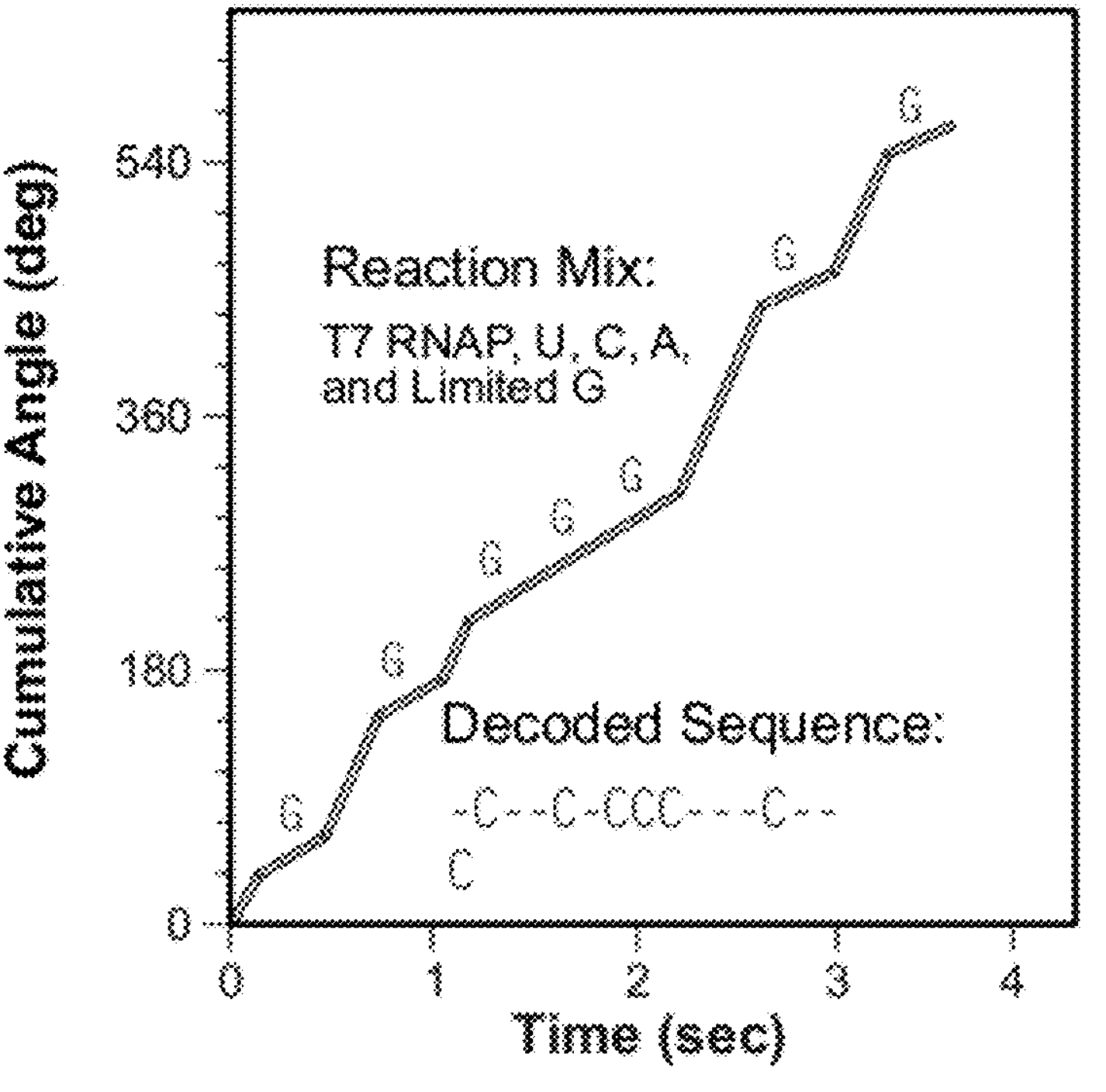
The patent mentions this of course, but focuses on a different approach. Here they supply all but one base in equal quantities. A 4th base is supplied in limit quantity. This means that every time the polymerase needs to incorporate the “limit quantity” base, it pauses and hangs around waiting for one to come along.
Assuming that the polymerase otherwise incorporates at a constant rate, you can use these pauses to detect where that base occurs on a strand.
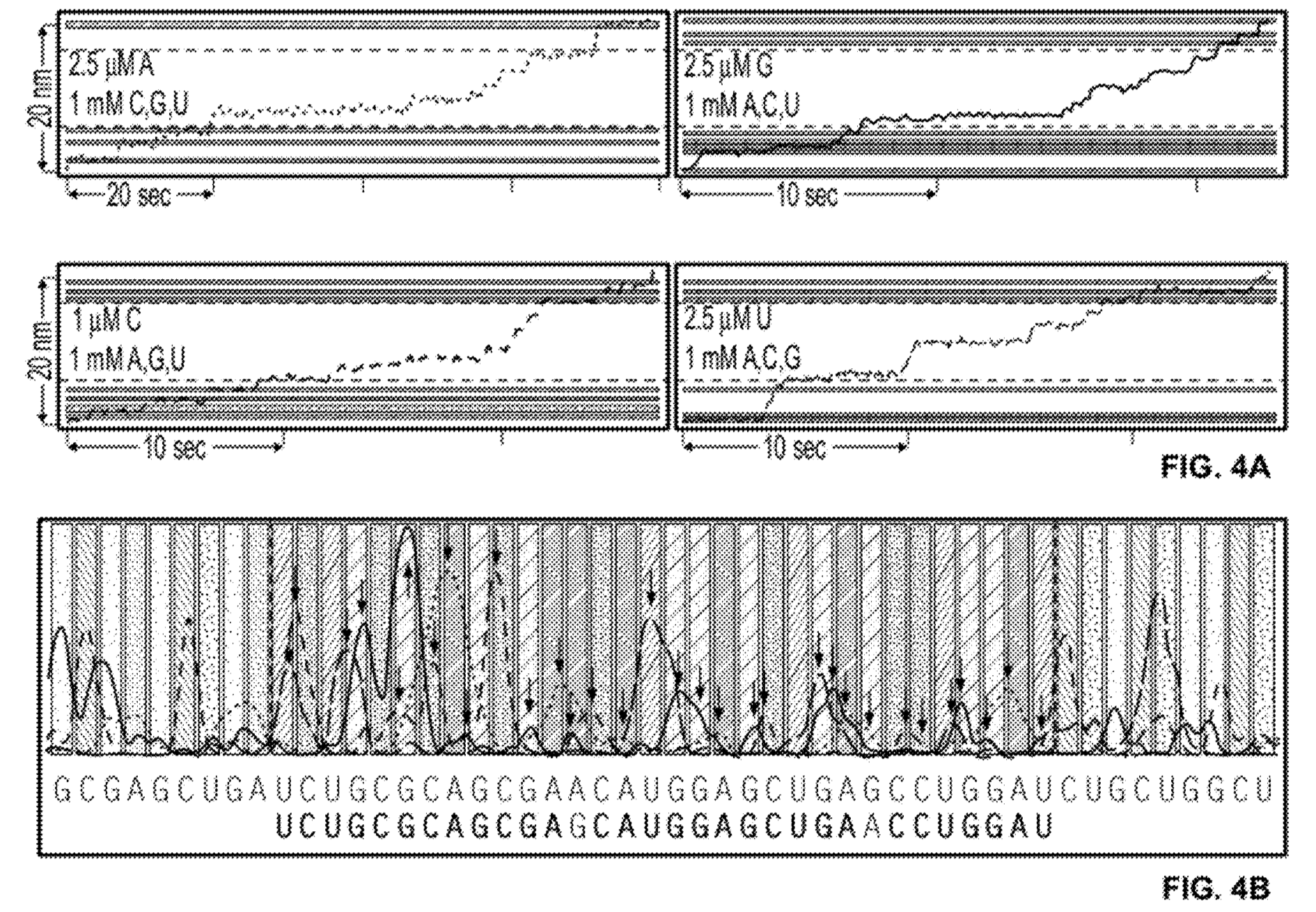
You then need to perform the sequencing experiment 4 times [6], limiting one base each time. The plot below, shows an example trace showing where slower incorporation indicates the incorporation of a “G”:
The patent also shows what appears to be a complete worked through real dataset (the text implies it’s real). In this example, 4 experiments are performed each reaction containing a limited quantity of one base. The rotation traces are captured and then combined to determine the template sequence:
The sequence contains a couple of homopolymers, I guess the incorporation process slows down for twice as long at these points. Overall, it’s difficult to tell exactly how well the process is working from the trace above (or get any idea of what the error rate might be). But if this is proof of concept real data, it looks pretty reasonable!
The approach seems attractive because it vastly reduces the number of cycles/washes that would need to be performed (just 4 versus 1 for every base position sequenced).
The Eve Biomedical approach has other advantages too, because the beads are big, the optical system should be relatively simple and cheap (commodity mobile phone CMOS sensors?). But there are disadvantages too. Because the sequencing is occurring in real-time sequencing is limited to a single field of view. This might make it harder to scale the platform.
Eve Biomedical have a second patent [1]. On my brief reading, this refers to using the same RNA polymerase, limited quality of one base system. However in this patent they suggest using it with a nanopore/nanostructure platform. The patent doesn’t seem to include real data/rig images.
The Eve Biomedical approach is really unique, and I’m not seen anyone else suggest using rotation to detect incorporation. While a very different approach, it reminds somewhat of the scheme Depixus have presented.
I’d love to see it played out a little more, if only because it’s so different.
Notes
[1] https://patents.google.com/patent/US20180010181A1
[1a] https://patents.google.com/patent/US20120214171A1
“As a consequence of transcription, the RNA polymerase exerts torque on the nucleic acid, which, in turn, manifests itself as rotation of a tag attached to the nucleic acid.”
“Such a method generaly includes contacting an RNA polymerase with a target nucleic acid molecule under sequencing conditions, detecting the rotational patern of the rotation tag, and repeating the contacting and detecting steps a plurality of times.”
“acid molecule comprises a rotation tag. The sequence of the target nucleic acid molecules is based, sequentialy, on the presence or absence of a change in the rotational patern in the presence of the at least one nucleoside triphosphate”
“FIG.3 are graphs showing two modes of nucleic acid sequencing described herein: Panel A shows an asynchronous, real-time “nucleotide patern’ sequencing strategy, where a limited concentration of a single nucleoside triphosphate (guanine(G) in this Panel) causes the polymerase to pause when incorporating G nucleotides in to the nascent Strand. Panel B shows asynchronous sequencing strategy, where a “base-by-base’ introduction of nucleoside triphosphates results in a continuous decoding of the nucleotide Sequence.”
“Rotation-dependent transcriptional sequencing relies upon transcription of target nucleic acid molecules by RNA polymerase. The RNA polymerase is immobilized on a solid surface, and a rotation tag is bound to the target nucleic acid molecules. During transcription, RNA polymerase establishes a transcription bubble in the template nucleic acid that contains within it an RNA:DNA hybrid of approximately 8 bases. As the RNA polymerase advances along the double-stranded nucleic acid template, it must unwind the helix at the leading edge of the bubble and reanneal the strands at the trailing edge. The torque produced as a result of the unwinding of the double-stranded helix results in rotation of the template nucleic acid relative to the RNA polymerase of about 36° per nucleotide incorporated. Therefore, when the RNA polymerase is immobilized on a solid surface and a rotation tag is attached to the template nucleic acid, the rotation of the template nucleic acid can be observed and is indicative of transcriptional activity (i.e., incorporation of a nucleoside triphosphate) by the enzyme.”
[2] https://www.genome.gov/27554929/
[3] ~1MUSD in SBIR grants: https://www.sbir.gov/sbirsearch/detail/671328
[4] Crunchbase lists a total of 7.7MUSD raised, from one disclosed investor DFJ. https://www.crunchbase.com/organization/eve-biomedical
[5] Here a figure from the patent which as I understand it shows magnetic beads going in and out of focus and the field strength is varied. This process isn’t used in sequencing, but shows how the beads/strands can be put under slight tension during the sequencing process.
[6] Strictly speaking 3 times, as you could obviously infer one of the bases as “not being any of the others”. You could also probably using the Cygnus type “mixtures of bases” error correction schemes with this approach.