Following on from my previous article on the BGI and list of sequencing companies this post contains my notes on Complete Genomics (a BGI acquisition). If you have any further insights on Complete, I’d love to hear them. Please email at new at sgenomics dot org.
Business
Complete Genomics was founded in 2006 to develop a DNA sequencing platform based on a sequencing-by-hybridisation (SBH) approach. One of the founders (Dr Drmanac) has a long history of academic work in SBH going back to the 1980s. The commercial history of Drmanac’s SBH work also starts before Complete with a company called Callida Genomics which was founded in 2001 as a subsidiary of HySeq. There are Callida Genomics patents referring to methods currently used by Complete [6], which are now assigned to them. Drmanac also cofounded Hyseq, and it seems likely that some SBH work went on there. There’s therefore a commercial history behind the Complete Genomics approach extending back 10 to 15 years now.
Complete Genomics IPO’d in 2010. Crunchbase notes that Complete raised a total of 143M USD [12], and were valued at 232.2M USD at IPO. They were then acquired in 2013 by the BGI (for 117.6M USD). The BGI seems to have transferred most technology development work to their Shenzhen site, cancelling new projects at the Complete Genomics Mountain View office [1] and making substantial staff cuts.
The BGI have however continued to develop the platform. Releasing sequencers for use in China under the BGISEQ brand. Competitively the Complete Genomics approach does not appear to have fared well against Illumina, but it remains and interesting technological approach.
Technology
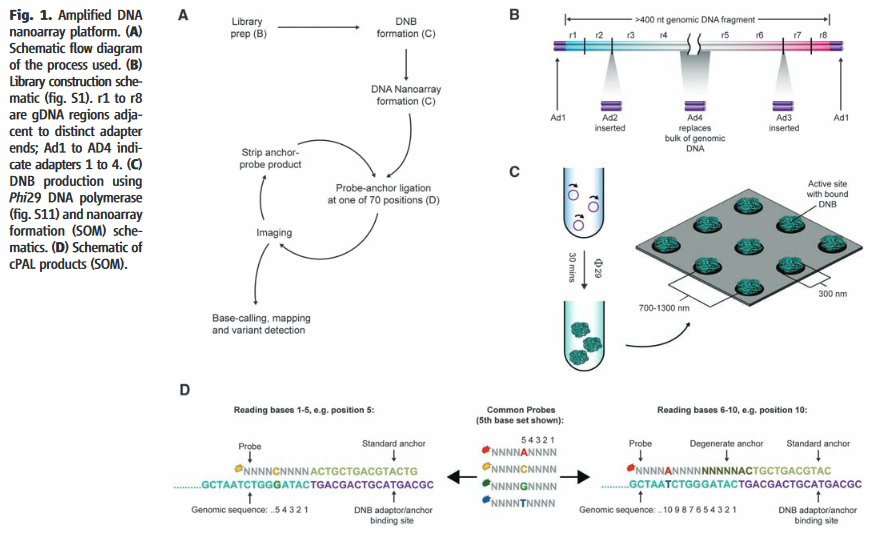
Technology overview from [3].
The Complete Genomics chemistry, as described in their 2010 paper, starts with the formation of nanoballs of DNA (DNBs) [8]. They appear to have a neat chemistry which allows them to form nanoballs in solution which are not entangled with neighboring DNBs [7]. This is in contrast to other platforms which either require amplification to be performed on beads, and/or in droplets (emulsion PCR) or on a surface (Illumina clusters, polonys).
The DNBs are flowed onto a substrate (flowcell). This flowcell is patterned with an array of aminosilane features. The DNBs only bind to these features, resulting in a regular array. From what I can tell, only a single DNB can bind to each site. Without this they could have overlapping nanoballs (which would unusable), in general such systems are limited to about a third of site containing a single read (with multiple occupancy sites being unusable). Potentially this gives them a density advantage over other DNA sequencing platforms.
With the DNBs arrayed on the chip sequencing can begin.
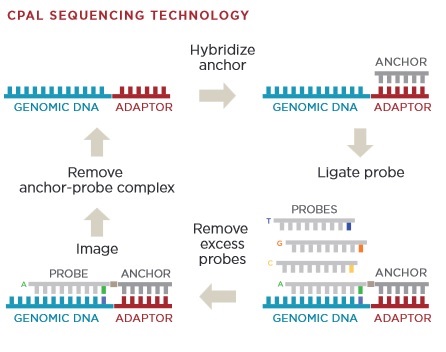
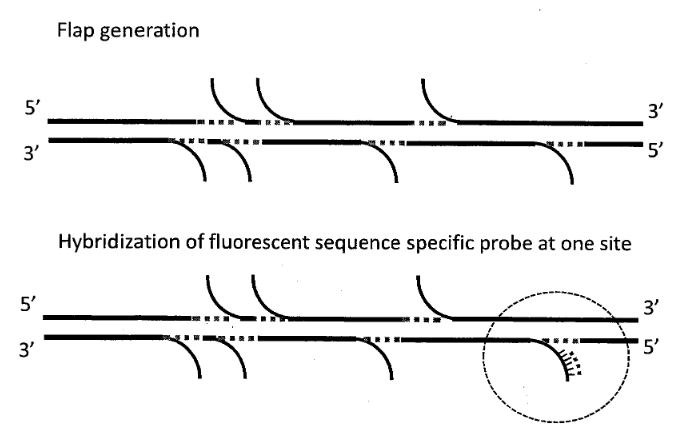
[9] Chemistry overview from Revolocity document.
The image above gives an overview of the cPAL sequencing chemistry used in Complete Genomics instruments. This hybridisation/ligation sequencing process is probably my least favorite part of the Complete Genomics system. The process uses fluorescently labelled degenerate 9mers with a single known position [11]. So for example, to interrogate the first position the probes NNNNNNNNA NNNNNNNNT NNNNNNNNG and NNNNNNNNC might be used, each labeled with a different dye. After these are flowed in, ligated and imaged they are then removed and the next set of probes comes in. These would then interrogate the next position NNNNNNNAN etc.
I’d guess 9mers are used because the stability of shorter oligos isn’t good enough. My understanding is that they only label the first 5 positions. They then have a process for creating extended anchor probes “by ligation of two anchor probes allows decoding of positions 6–10 adjacent to the adaptor” [10]. This results in 10mer reads. They perform a number of 10mer reads at different adaptor sites and merge everything together. From memories of early datasets, reads have the potential for gaps because of this.
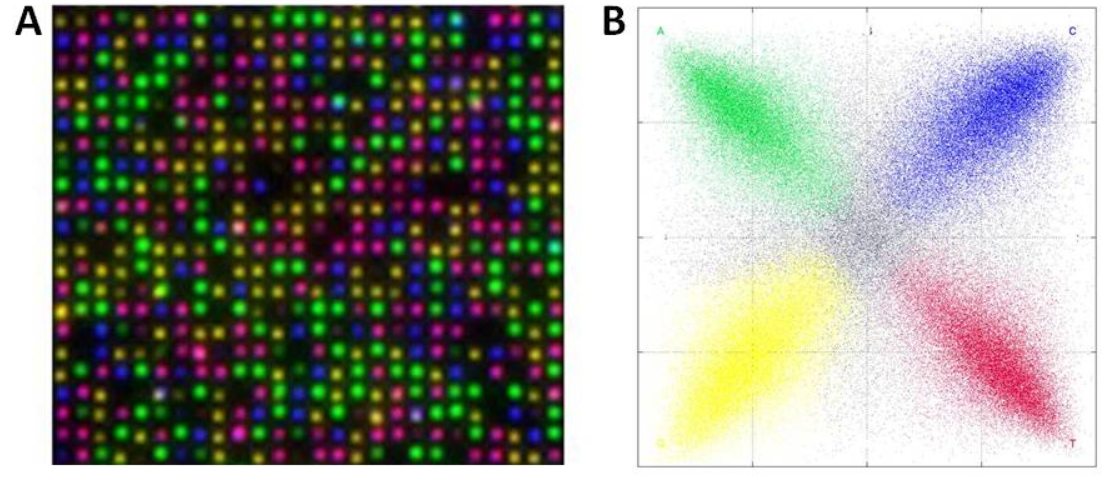
Un-captioned image from [3] Supplementary info. I assume the image on the left is the combined output of 4 images of a single cycle. Image on the right possibly represents crosstalk between dyes.
The process is complex and potentially error prone. However there is one possible advantage over the Illumina SBS approach. That is that each sequencing cycle in the cPAL system resets the template by removing the probes. In Illumina sequencing there is the potential for accumulated error (phasing error) as templates get out of sync (which ultimately limits read length). This does not exist here.
I’ve not looked at the original Complete Genomics raw data (I don’t believe any was released?). But I’d guess there is a potentially high raw read error rate (due to non-specific hybridisation among other things). It’s unlikely standard short read aligners would work well with Complete Genomics reads (being short and possibly containing gaps). For this, and other reasons Complete Genomics only ran a service business for many years. As I recall, they would only process human genomes, and delivered called SNPs to the customer rather than read data itself.
While the Complete Genomics systems were only used in house for a long time, since the acquisition of Complete by the BGI a line of commercial sequencers has been released under the BGISEQ brand. The BGISEQ-500 spec sheet suggests they are now generating 50bp reads [11] (the approach seems to be similar). Another instrument series, the MGISEQ has also been announced, which boasts a 100bp read length, however little information is available. It looks like fastq files may now be generated. Not much data seems to have made its way into the various public archives, but there is one report that suggests BGISEQ data looks pretty reasonable for SNP calling. Update: It’s not clear that MGI are pursuing an SBS approach, not SBH as used by Complete.
The approach is technologically interesting, but it’s difficult to see how it can complete directly with Illumina. However, it seems likely the BGISEQ instruments have a significant cost advantage at least in China.
Notes
[1] https://www.genomeweb.com/sequencing-technology/bgi-halts-revolocity-launch-cuts-complete-genomics-staff-part-strategic-shift
[2] Callida Genomics, Inc: http://www.evaluategroup.com/Universal/View.aspx?type=Story&id=13022
[3] http://science.sciencemag.org/content/327/5961/78
[4] Revolocity Video https://www.youtube.com/watch?v=WuS_RY8Zy38
[5] https://www.youtube.com/watch?v=DfaMOTcwcjs
[6] Callida Genomics Patent, referring to nanoball approach: https://patents.google.com/patent/US8440397
[7] “Short palindromes in the adaptors promote coiling of ssDNA concatamers via reversible intra‐molecular hybridization into compact ~300 nm DNBs, thereby avoiding entanglement with neighboring replicons” Science 2010 [3], supplementary info. Documentation for the now cancelled Revolocity states that “>95% occupancy of flow cell spots occupied by a single DNB”. http://www.completegenomics.com/documents/revolocity-tech-overview.pdf
[8] Using a controlled, synchronized synthesis, we obtained hundreds of tandem copies of the sequencing substrate in palindrome-promoted coils of single-stranded DNA, referred to as DNA nanoballs (DNBs).
[9] http://www.completegenomics.com/documents/revolocity-tech-overview.pdf
[10] The process is described well here: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3472021/
[11] https://www.bgi.com/us/wp-content/uploads/sites/2/2017/04/BGISEQ-500-ChIP-Service-Overview_linear.pdf
[12] Complete Genomics investors appear to include:
Enterprise Partners, OVP Venture Partners, Highland Capital Partners, Orbimed, Essex Woodlands Health, Prospect Venture Partners, Sands Capital Management, and ATEL Capital Group