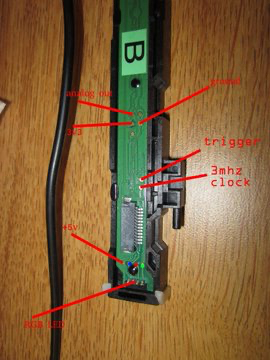
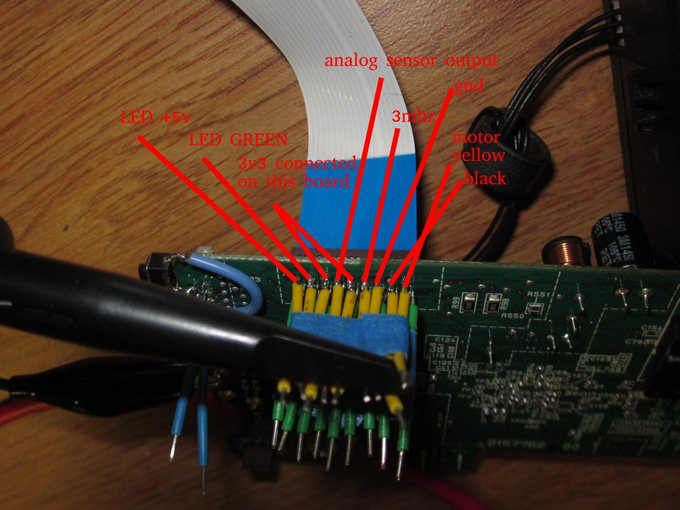
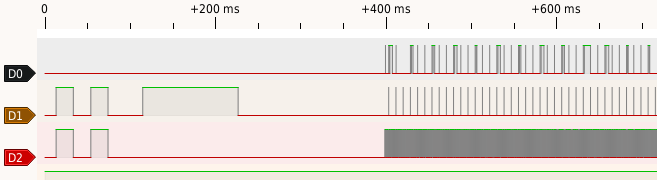
|
#include <Arduino.h> |
|
#include <ports.h> |
|
#include <base.h> |
|
#include <DirectIO.h> |
|
|
|
#include <Adafruit_NeoPixel.h> |
|
#include <TimerOne.h> |
|
|
|
#define CLKPIN TIMER1_A_PIN |
|
#define TRGPIN 4 |
|
#define DEBUGPIN 6 |
|
#define FIRSTFIELD 7 |
|
|
|
#ifndef cbi |
|
#define cbi(sfr, bit) (_SFR_BYTE(sfr) &= ~_BV(bit)) |
|
#endif |
|
#ifndef sbi |
|
#define sbi(sfr, bit) (_SFR_BYTE(sfr) |= _BV(bit)) |
|
#endif |
|
|
|
Output<TRGPIN> trg; |
|
Output<CLKPIN> clk; |
|
Output<6> dbg; |
|
Output<FIRSTFIELD> ff; |
|
|
|
#define NUMPIXELS 8 |
|
#define NEO_PIN 8 |
|
Adafruit_NeoPixel pixels = Adafruit_NeoPixel(NUMPIXELS, NEO_PIN, NEO_RGB + NEO_KHZ800); |
|
|
|
const uint8_t PROGMEM gamma8[] = { |
|
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, |
|
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1, 1, |
|
1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 2, 2, |
|
2, 3, 3, 3, 3, 3, 3, 3, 4, 4, 4, 4, 4, 5, 5, 5, |
|
5, 6, 6, 6, 6, 7, 7, 7, 7, 8, 8, 8, 9, 9, 9, 10, |
|
10, 10, 11, 11, 11, 12, 12, 13, 13, 13, 14, 14, 15, 15, 16, 16, |
|
17, 17, 18, 18, 19, 19, 20, 20, 21, 21, 22, 22, 23, 24, 24, 25, |
|
25, 26, 27, 27, 28, 29, 29, 30, 31, 32, 32, 33, 34, 35, 35, 36, |
|
37, 38, 39, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 50, |
|
51, 52, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 66, 67, 68, |
|
69, 70, 72, 73, 74, 75, 77, 78, 79, 81, 82, 83, 85, 86, 87, 89, |
|
90, 92, 93, 95, 96, 98, 99,101,102,104,105,107,109,110,112,114, |
|
115,117,119,120,122,124,126,127,129,131,133,135,137,138,140,142, |
|
144,146,148,150,152,154,156,158,160,162,164,167,169,171,173,175, |
|
177,180,182,184,186,189,191,193,196,198,200,203,205,208,210,213, |
|
215,218,220,223,225,228,231,233,236,239,241,244,247,249,252,255 }; |
|
|
|
void setup() { |
|
analogReference(EXTERNAL); |
|
pinMode(CLKPIN, OUTPUT); |
|
//pinMode(DEBUGPIN, OUTPUT); |
|
|
|
Timer1.initialize(1); // ~1mhz |
|
Timer1.pwm(CLKPIN, 512); // 50% duty cycle |
|
Timer1.stop(); |
|
pinMode(TRGPIN, OUTPUT); |
|
pinMode(A0, INPUT); |
|
pinMode(ledToPin(0), OUTPUT); |
|
pinMode(ledToPin(1), OUTPUT); |
|
pinMode(ledToPin(2), OUTPUT); |
|
Serial.begin(115200); |
|
cbi(ADCSRA, ADPS0); |
|
cbi(ADCSRA, ADPS1); |
|
cbi(ADCSRA, ADPS2); |
|
sbi(ADCSRA, ADPS1); |
|
|
|
// Serial.print("prescaler = "); |
|
// Serial.println(ADCSRA & 0xE0); |
|
pixels.begin(); |
|
} |
|
|
|
int avgRead(int port, int samples) |
|
{ |
|
int i; |
|
long reading = 0; |
|
for(i = 0; i < samples; i++) |
|
{ |
|
reading += analogRead(port); |
|
} |
|
|
|
reading /= samples; |
|
|
|
return reading; |
|
} |
|
|
|
void clockCycle(int pin, int count) |
|
{ |
|
while(count–) |
|
{ |
|
digitalWrite(pin, HIGH); |
|
digitalWrite(pin, LOW); |
|
} |
|
} |
|
|
|
int ledToPin(int color) |
|
{ |
|
switch(color) |
|
{ |
|
case 0: // red |
|
return 2; |
|
break; |
|
|
|
case 1: // green |
|
return 3; |
|
break; |
|
|
|
case 2: // blue |
|
return 5; |
|
break; |
|
} |
|
} |
|
void ledOn(int color) |
|
{ |
|
digitalWrite(ledToPin(color), LOW); |
|
} |
|
|
|
void ledOff(int color) |
|
{ |
|
digitalWrite(ledToPin(color), HIGH); |
|
} |
|
|
|
void ledsOn() |
|
{ |
|
for(int i = 0; i <= 2; i++) |
|
ledOn(i); |
|
} |
|
|
|
void ledsOff() |
|
{ |
|
for(int i = 0; i <= 2; i++) |
|
ledOff(i); |
|
} |
|
|
|
// returns n samples from start of readback to end of readback |
|
void readLine(byte* output, int samples) |
|
{ |
|
int zeroLevel; |
|
int blackLevel; |
|
int clocks; |
|
int i; |
|
|
|
int samplePos = 2593 / samples; |
|
int currSample = 0; |
|
|
|
shiftOut(TRGPIN, CLKPIN, MSBFIRST, 0xF0); // trigger pulse and slow clock |
|
|
|
// lock step through front porch with slow clock in order to capture reference levels without unpredictable jitter |
|
clockCycle(CLKPIN, 4); |
|
|
|
// sample black level |
|
for(i=0; i < 3; i++) |
|
{ |
|
clocks=5; while(clocks–) |
|
{ |
|
clk=1; |
|
clk=0; |
|
} |
|
blackLevel += avgRead(A0, 3); |
|
} |
|
|
|
blackLevel /= 3; |
|
// Serial.print("blackLevel = "); |
|
// Serial.println(blackLevel); |
|
|
|
// 65 more clock cycles until analog goodies |
|
clocks = 65; while(clocks–) |
|
{ |
|
clk=1; |
|
clk=0; |
|
} |
|
|
|
#define ANALOG_CLOCKS (12 * 216 + 1) |
|
|
|
int stepSpacing = ANALOG_CLOCKS / samples; |
|
int nextSample = 0; |
|
|
|
// go through analog bins |
|
for(i = 0; i < 12 * 216 + 1; i++) |
|
{ |
|
clk = 1; |
|
|
|
if(nextSample– == 0) |
|
{ |
|
dbg=1; |
|
nextSample = stepSpacing; |
|
// output[currSample++] = map(avgRead(A0, 5), blackLevel, 1024, 0, 255); |
|
output[currSample++] = map(analogRead(A0), blackLevel, 1024, 0, 255); |
|
dbg=0; |
|
} |
|
clk = 0; |
|
} |
|
|
|
#if 0 |
|
for(i = 0; i < 2593; i++) |
|
{ |
|
clk =1; |
|
clk =0; |
|
} |
|
#endif |
|
|
|
} |
|
|
|
void loop() { |
|
int led = 0; |
|
byte samples[64]; |
|
byte redSamples[NUMPIXELS]; |
|
byte greenSamples[NUMPIXELS]; |
|
byte blueSamples[NUMPIXELS]; |
|
int i; |
|
|
|
ff=1; |
|
ff=0; |
|
|
|
#if 1 |
|
ledOn(0); |
|
readLine(redSamples, NUMPIXELS); |
|
ledsOff(); |
|
|
|
ledOn(1); |
|
readLine(greenSamples, NUMPIXELS); |
|
ledsOff(); |
|
|
|
ledOn(2); |
|
readLine(blueSamples, NUMPIXELS); |
|
ledsOff(); |
|
#endif |
|
#if 0 |
|
for(led = 0; led < 0; led++){ |
|
ledOn(led); |
|
readLine(samples, 4); |
|
ledOff(led); |
|
} |
|
ledsOff(); |
|
|
|
#endif |
|
|
|
for(i = 0; i < NUMPIXELS; i++) |
|
{ |
|
#if 0 |
|
Serial.print("sample["); |
|
Serial.print(i); |
|
Serial.println("] = "); |
|
Serial.print("red "); |
|
Serial.println(redSamples[i]); |
|
Serial.print("green "); |
|
Serial.println(greenSamples[i]); |
|
Serial.print("blue "); |
|
Serial.println(blueSamples[i]); |
|
#endif |
|
|
|
|
|
pixels.setPixelColor(i, pixels.Color(pgm_read_byte(&gamma8[greenSamples[i]]), pgm_read_byte(&gamma8[redSamples[i]]), pgm_read_byte(&gamma8[blueSamples[i]]))); |
|
|
|
// pixels.setPixelColor(i, pixels.Color(0, 0, 0)); |
|
// pixels.setPixelColor(i, pixels.Color(redSamples[i], greenSamples[i], blueSamples[i])); |
|
// pixels.setPixelColor(i, pixels.Color(pgm_read_byte(&gamma8[redSamples[i]]), pgm_read_byte(&gamma8[greenSamples[i]]), pgm_read_byte(&gamma8[blueSamples[i]]))); |
|
// pixels.setPixelColor(i, pixels.Color(pgm_read_byte(&gamma8[redSamples[i]]), pgm_read_byte(&gamma8[redSamples[i]]), pgm_read_byte(&gamma8[redSamples[i]]))); |
|
|
|
pixels.show(); // This sends the updated pixel color to the hardware. |
|
|
|
|
|
} |
|
// delay(100); |
|
//delayMicroseconds(100); |
|
} |
|
|
|
void old_loop() { |
|
int bin; |
|
int i; |
|
int samples[12]; |
|
int clocks; |
|
int zeroLevel; |
|
int blackLevel; |
|
static int led=0; |
|
//shiftOut(TRGPIN, CLKPIN, MSBFIRST, 0); |
|
dbg=1; |
|
zeroLevel = avgRead(A0, 5); |
|
// zeroLevel = analogRead(A0); |
|
dbg=0; |
|
|
|
dbg=1; |
|
shiftOut(TRGPIN, CLKPIN, MSBFIRST, 0xF0); // trigger pulse and slow clock |
|
dbg=0; |
|
|
|
// lock step through front porch with slow clock in order to capture reference levels without unpredictable jitter |
|
dbg=1; |
|
clockCycle(CLKPIN, 4); |
|
dbg=0; |
|
|
|
dbg=1; |
|
for(i=0; i < 3; i++) |
|
{ |
|
clocks=5; while(clocks–) |
|
{ |
|
clk=1; |
|
clk=0; |
|
} |
|
blackLevel += avgRead(A0, 3); |
|
} |
|
|
|
blackLevel /= 4; |
|
// Serial.print("blackLevel = "); |
|
// Serial.println(blackLevel); |
|
dbg=0; |
|
|
|
if(led == 0){ |
|
ff = 1; |
|
ff = 0; |
|
} |
|
ledOn(led); |
|
//ledsOn(); |
|
dbg=1; |
|
clocks = 40; while(clocks–) |
|
{ |
|
clk=1; |
|
clk=0; |
|
} |
|
dbg=0; |
|
|
|
for(i = 0; i < 350*8; i++) |
|
{ |
|
clk =1; |
|
clk =0; |
|
} |
|
dbg =1; |
|
|
|
dbg=0; |
|
// Timer1.pwm(CLKPIN, 512); // fast clock |
|
/* dbg=1; |
|
for(i = 0; i < 300; i++) |
|
{ |
|
samples[i] = analogRead(A0); |
|
//delayMicroseconds(2000); |
|
} |
|
dbg=0; |
|
*/ |
|
// sample front porch reference level 2 clocks later, for 80 hs clocks |
|
// sample bins 83 clocks after fast start |
|
// bins seem to be 215 clocks wide, 12 bins, 1 clock per pixel |
|
|
|
//delayMicroseconds(1); |
|
#if 0 |
|
dbg=1; |
|
noInterrupts(); |
|
while(analogRead(A0) -5 < zeroLevel) |
|
; |
|
interrupts(); |
|
// wait for voltage to increase to follow clock jitter |
|
dbg=0; |
|
#endif |
|
|
|
|
|
dbg = 1; |
|
blackLevel = avgRead(A0, 5); |
|
// blackLevel = analogRead(A0); |
|
dbg = 0; |
|
|
|
#if 0 |
|
noInterrupts(); |
|
while(analogRead(A0) -10< blackLevel) |
|
{ |
|
dbg=1; |
|
delayMicroseconds(0); |
|
dbg=0; |
|
} |
|
; |
|
interrupts(); |
|
|
|
#endif |
|
|
|
dbg = 1; |
|
delayMicroseconds(1); |
|
dbg = 0; |
|
/* |
|
delayMicroseconds(100); |
|
dbg=1; |
|
bin= analogRead(A0); |
|
dbg=0; |
|
*/ |
|
|
|
// delayMicroseconds(2670); |
|
Timer1.stop(); |
|
digitalWrite(CLKPIN, LOW); |
|
/* |
|
Serial.print("zeroLevel = "); |
|
Serial.println(zeroLevel); |
|
|
|
Serial.print("blackLevel = "); |
|
Serial.println(blackLevel); |
|
/* Serial.print("bin = "); |
|
Serial.println(bin); |
|
*/ |
|
|
|
//delay(50); |
|
#if 1 |
|
ledOff(led); |
|
// switch LED |
|
if(++led > 2) |
|
led = 0; |
|
#endif |
|
//ledsOff(); |
|
|
|
// Serial.println(led); |
|
// Serial.print("normalized = "); |
|
// Serial.println(map(bin – blackLevel, zeroLevel, 1024, 0, 255)); |
|
//delay(10); // inter line delay |
|
delayMicroseconds(100); |
|
} |