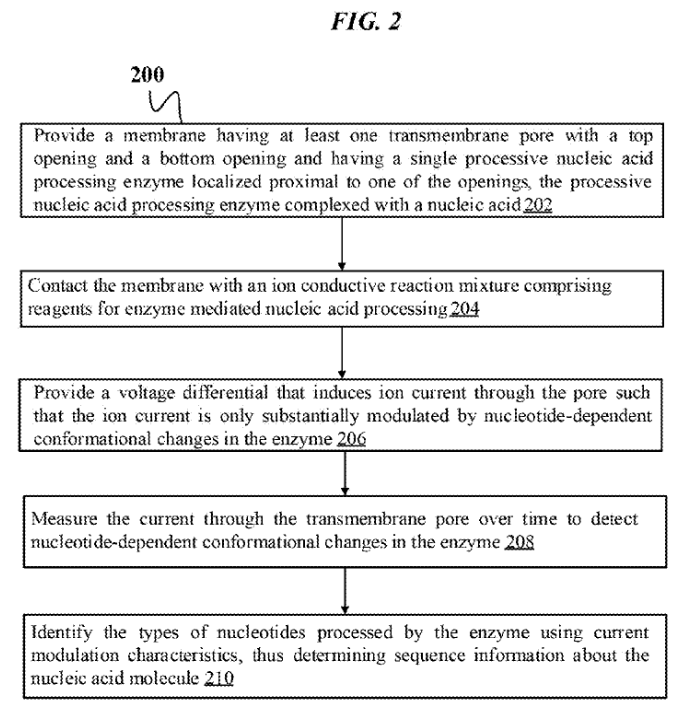
Stratos’ Other Approach…
Reading through some of Stratos’ more recent patents I came across a non-expandomer sequencing approach which I found quite interesting. The patent shows that Stratos had been thinking about other approaches to nanopore based sequencing. This suggests that the Roche acquisition may not just have been for their expandomer IP…
The Approach
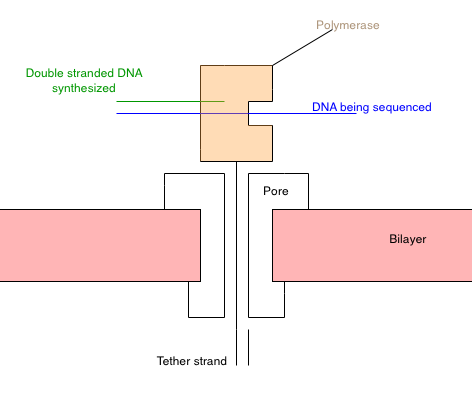
My understanding of the approach from [1] is summarized in the schematic above. Essentially you construct a polymerase [2] that has a “tether” attached to it. The tether is composed of a PEG repeat, which threads through a nanopore [3]. The PEG region has a short oligo on the end. Once it’s threaded through the pore, another oligo can be hybridized to it. This secures the tether in the pore.
With polymerase-tether complex in the pore, the system should look something like the diagram above. The polymerase is secured on the top of the pore. Due to its charge, I guess the polymerase would be pulled toward the pore, but it’s too big to pass through.
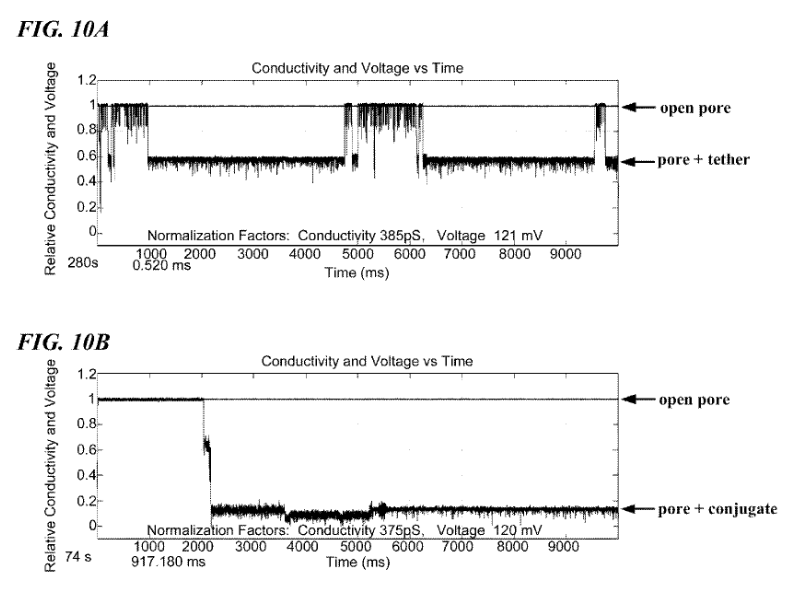
As in a standard Ionic/Protein nanopore setup, there’s a bias voltage, and ionic current passing through the pore. The polymerase is now blocking the pore. This causes a significant reduction in current flow. One of the figures in the patent illustrates this (and looks like experimental data):
Now, a template to be sequenced is introduced. Single stranded DNA enters the polymerase, and synthesis of the complementary strand occurs. However, the template DNA doesn’t interact with the pore directly.
The idea is that as the polymerase incorporates bases it will undergo confirmational changes. The patent suggests that these confirmational changes can be up to about 1nm.
The idea here is that different confirmational changes will cause the polymerase to block the pore with varying efficiency. So, for example when incorporating a base it might go through a series of confirmational changes, which result in less current flow, then more current flow, then back to baseline.
Ideally, each base would induce a distinct conformational change, and current blockage. The current trace can then be used to infer the sequence of the DNA template.
However, if you’re only able to detect incorporation/non-incorporation then nucleotides could be sequentially introduced.
The approach is somewhat reminiscent of that proposed by Roswell. Both of which seem to be trying to detect confirmational changes in the polymerase. In comparison to the Roswell approach, this seems like a simpler setup. The polymerase is also closer to the “sensor” which might make detecting confirmation changes easier.
Contrasting this with other ionic nanopore approaches, it’s kind of nice that the strand doesn’t go through the pore. Ideally in the Stratos approach there would be at most 4 different signal types, one for each base. Rather than the signal being some combination of all the nucleotides currently in the pore. Theoretically this could result in lower error rates.
Overall the idea seems at least plausible, and I’ll be curious to see how it plays out.
References and Notes
[1] http://www.freepatentsonline.com/20170159115.pdf
[2] A number of enzymes that process DNA could work (for example an exonuclease). And the patent states this, but a polymerase seems the most likely option.
[3] They talk about and show various protein nanopores. But I imagine a solid state nanopore may also be viable, particularly as the dimensions of the pore maybe slightly less critical in this approach.
[4] “The tethers were constructed of three domains (i.e., “segments”): 1) a polyethylene glycol (PEG) repeat region, located proximal to the polymerase and designed to span the nanopore channel; 2) a short oligonucleotide, designed to hybridize to a single-stranded oligonucleotide on the opposite side of the nanopore relative to the polymerase to anchor the assembly; and 3) a negatively charged phosphoramidite tail, located most distal to the polymerase and designed to facilitate threading of the tether through the nanopore. FIG. 9 is a SDS/PAGE gel that shows the size of the unmodified KF polymerase (lane 1), the KF-tether 1 conjugate (lane 2) and the KF-tether 2 conjugate (lane 3). As expected, the conjugates show an increase in mass compared to the unmodified polymerase.”
[5] “When this polymerase complexes with a nucleotide that is the complement to the template base in the next extension position the polymerase reconfigures into what is referred to in the art as a “closed” conformation. At a more detailed structural level, the transition from the open to closed conformation is characterized by relative movement within the polymerase resulting in the “thumb” domain and “fingers” domain being closer to each other. In the open conformation the thumb domain is further from the fingers domain, akin to the opening and closing of the palm of a hand. In various polymerases, the distance between the tip of the finger and the thumb can change up to 10 angstroms between the “open” and “closed” conformations. The distance between the tip of the finger and the rest of the protein domains can also change up to 10 Angstroms. It will be understood that this change will be exploited in a method set forth herein.”
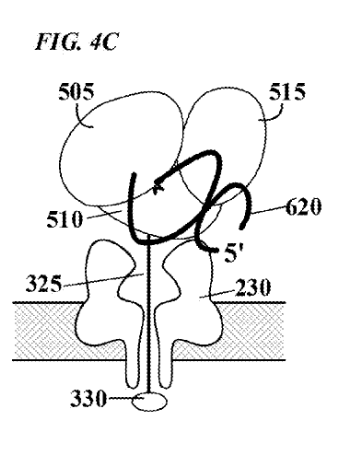
[6] This figure in the patent is supposed to show the threading/polymerase. But I find it a bit confusing.
Other quotes….
“The present disclosure relates to methods and constructs for single molecule electronic sequencing of template nucleic acids. The constructs are molecular sensor complexes which comprise a processive nucleic acid processing enzyme localized to a nanopore. Conformational changes in the enzyme induced by single nucleic acid processing events are transduced into electric signals by the nanopore, which are used to identify individual nucleotides. The methods can include the steps of providing a membrane with the nanopore and the enzyme complexed with a template nucleic acid localized proximal to an opening in the pore, contacting the enzyme with an ion conductive reaction mixture including the reagents required for nucleic acid processing, providing a voltage drop across the pore that induces ion current through the pore that is modulated by conformational changes in the enzyme, measuring current through the pore over time to detect nucleotide-dependent conformational changes in the enzyme, and identifying the type of nucleotide processed by the enzyme using current modulation characteristics, thus determining sequencing information about the nucleic acid molecule.”
“The polymerase is secured to the pore by hybridizing a short oligonucleotide anchor to the tether construct on the distal side of the nanopore.”
“These results indicate that a tether and a tether-polymerase conjugate can be anchored to a nanopore and, moreover, that the resulting complex can generate reproducible electrical signals. Polymerase-nanopore complexes are thus capable of modulating current flow through the pore and show promise as useful sensors to transduct mechanical events into electrical signals.”
“One or more of the transitions that a polymerase undergoes when adding a nucleotide to a nucleic acid can be detected using a molecular sensor complex as described herein.”
“FIG. 4C depicts the polymerase in a second, ™, “closed” configuration, which is induced, e.g., by binding of incoming nucleotide 605 to form a correct base pair with the template nucleic acid. In this second configuration, the degree to which the enzyme physically occludes the pore is reduced, and consequently the flow of current through the pore will increase. Such modulation of current flow generates an electronic signal specific for nucleotide species”
“In one embodiment, each of the four nucleotides induces a different polymerase conformation, as illustrated in FIG. 4C. . The movement of the polymerase during the incorporation of a nucleotide will modulate the ion current through the pore in a characteristic and reproducible manner, generating a signature electric signal.”
“In another embodiment, the average amplitude of the current modulation doesn’t change, but rather the noise in the current modulation changes as a single nucleotide is bound and incorporated. In yet another embodiment, the current modulation system only indicates an incorporation event but does not discriminate the base type. In this embodiment, the sequence information about a nucleic acid is obtained by sequentially flooding the senor complex with one of four reaction mixtures containing one of the four nucleotides and detecting the presence or absence of an electric signal.”
“For proper function of the molecular sensor complexes of the present invention, it is necessary that the enzyme be stably localized to the pore in sufficiently close proximity to reliably influence, or modulate, current flow through the pore. Several alternative localization and/or attachment structures or compositions are contemplated by the present invention, some which are illustrated schematically in FIGS. 5-8. FIG. 5A depicts one embodiment in which enzyme 500 is localized to pore 220 by covalent attachment to tethering structure 325, herein referred to simply as a “tether”. Tethers may be designed to thread through the lumen of the pore, from one side of membrane 100 to the other. Tethers may comprise one or more structural domains, or “segments”, designed to perform one or more functions.”