Caerus Molecular Diagnostics
Business
There’s not much business information available on Caerus, they’ve received ~1.2M USD in SBIR grants [1] most recently in 2014. I can currently only find one employee on LinkedIn (one of the founders, Javier Farinas), though I can find 3 other previous employees (including the co-founder, Andrea Chow). Caerus received their first grant in 2010, and I would guess they would likely need to raise soon to sustain R&D work on their platform.
Technology
Caerus originally proposed using a method they called Millikan sequencing. This was a method measuring the charge of a strand as nucleotides were incorporated. They published a proof on concept of this system in 2015 [2], however they also announced that they were abandoning the approach around this time [3].
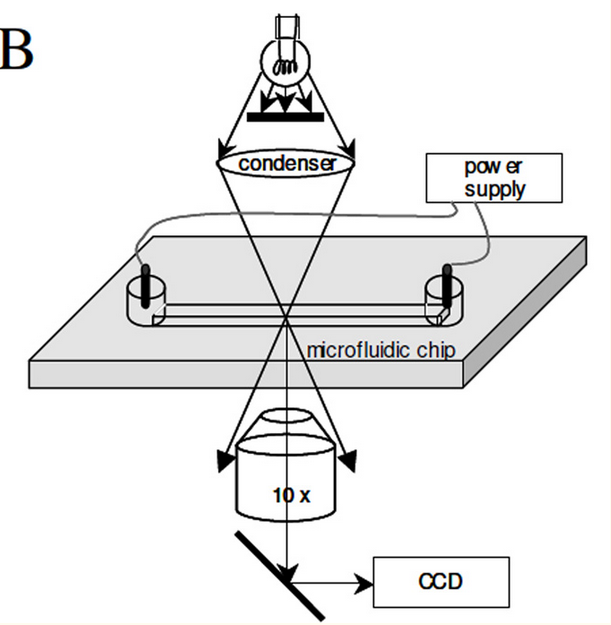
The Millikan sequencing approach was pretty interesting. In the proof of concept they present a system where a bead covered with multiple DNA templates is suspended between two electrodes.
They apply an AC bias voltage and can see the bead moving under the changing electric field (the DNA being negatively charged). The proof on concept uses sequencing by synthesis. As bases are incorporated the charge on the bead changes and these incorporations can be detect by changes in the motion of the bead (they use a velocity measurement).
It’s pretty neat that they could use natural, unlabeled nucleotides (which could potentially lead to longer reads). But they have apparently abandoned the approach. Because they used amplified DNA, they would be subject to the same phasing errors that limit read length on other platform. Ion torrent also used unlabeled bases and I guess they figured the competitive advantage was too small. Having an optical sequencing technology that uses unlabeled bases is kind of neat however (but I guess also similar to 454s platform).
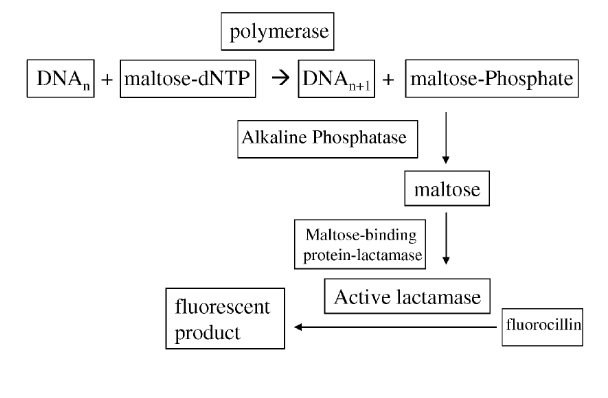
After abandoning the Millikan approach, they started working on something called “activator sequencing”. This method is covered in their 2014 patent [4].
The patent states “the methods use an enzyme to covert a product produced from a sequencing reaction into many copies of a readily detectable report molecule”. Essentially what they appear to be suggesting is single molecule sequencing [5] system where incorporation activates an enzyme which in turn creates many copies of a detectable (e.g. fluorescent) molecule.
It seems like an interesting idea, but at the single molecule it’s possible that engineering efficient enzyme activation might be problematic. It will be interesting to see how things progress!
Notes
[1] https://www.sbir.gov/sbirsearch/detail/12662
[2] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4560655/
[3] https://www.genomeweb.com/sequencing-technology/caerus-molecular-explores-activator-sequencing-long-single-molecule-reads-low
“For amplified DNA, they did show proof-of-principle of sequencing, which they published last month in Analytical Biochemistry. However, it did not make sense to develop that approach commercially. “It was pretty evident that we were not being able to catch up to the rest of the field using that method,” Farinas recalled, noting that Ion Torrent came out with the PGM around the same time, which offered many of the same benefits, including label-free sequencing, that Millikan sequencing promised. “Basically, they had a commercial product and we had some preliminary data,” he said. ”
[4] http://www.freepatentsonline.com/20160068902.pdf
All Patents:
Click to access 20120220486.pdf
http://www.freepatentsonline.com/y2016/0068902.html
http://www.freepatentsonline.com/y2010/0112588.html
http://www.freepatentsonline.com/y2011/0059864.html
[5] The patent also suggests they the scheme could be used with colonies (clusters).