Inside the Lucira Check It COVID-19 Test
This post was previously published on the substack.
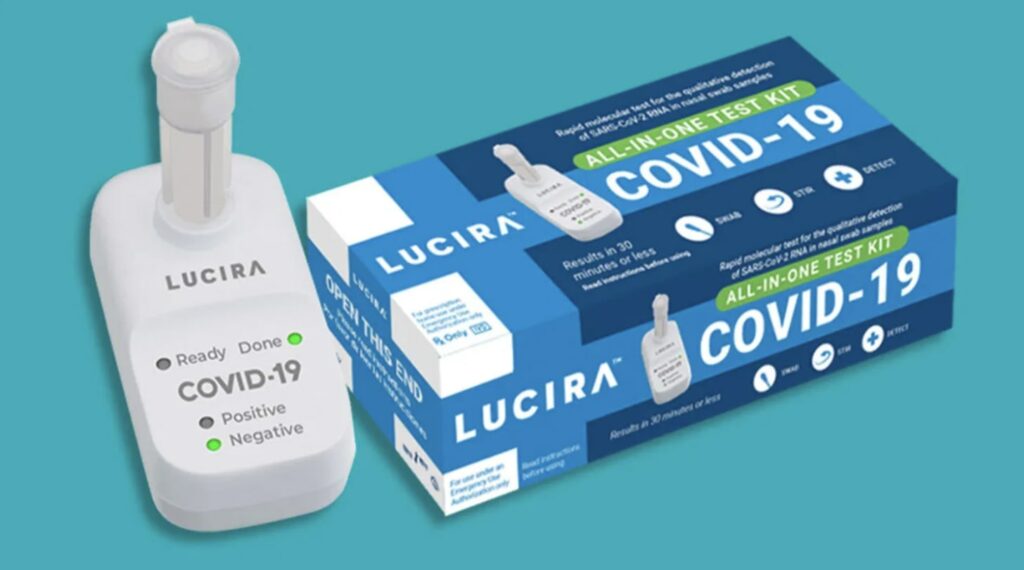
The Lucira COVID-19 test is an “at-home” LAMP qPCR-like test. Essentially you receive disposable molecular test device, with embedded reagents for $55 which you use once and then throw away.
I was curious to better understand the instrument and went hunting for teardown pictures and patents. Luckily Brad Ackerman on pulled one apart and posted pictures on twitter. Some of these are included here with permission from Brad. The teardown pictures also closely match one of Lucira’s patents which gives us another source of information. The patent has some wonderful figures, and gives us a really nice exploded view of the device:
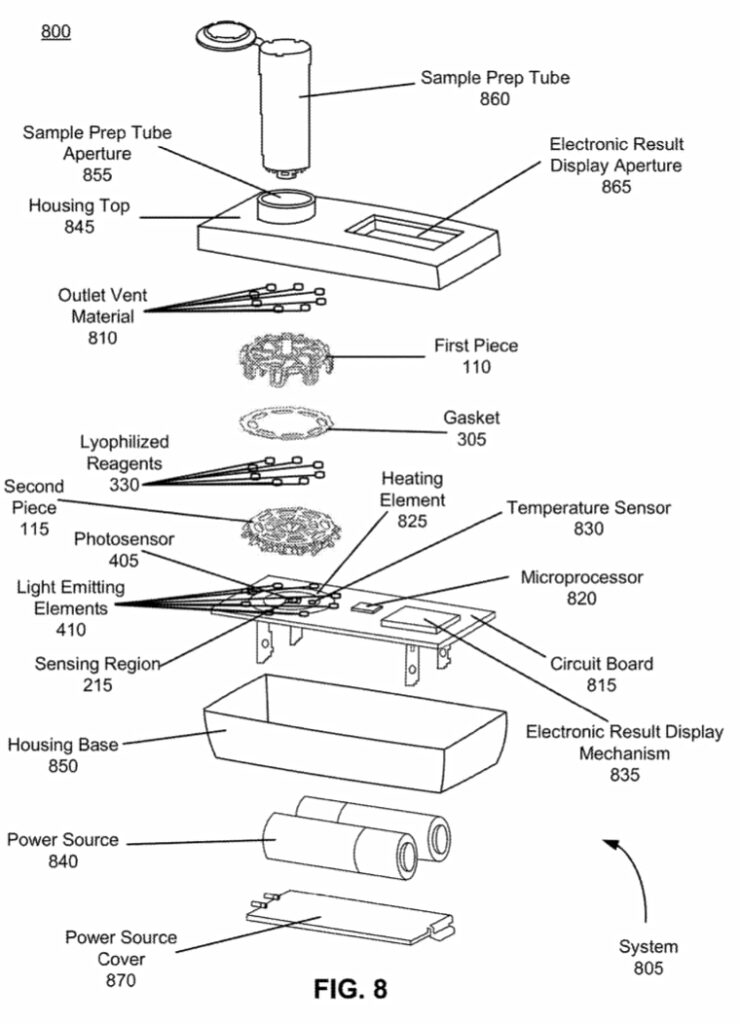
This really gives us a great overview of the instrument. We can see a single main PCB with contains LEDs, a photosensor, heating element, temperature sensor and microcontroller. The heating element is a simple PCB trace used for resistive heating. This is required get the reaction to ~60C as required for LAMP. Critically, for LAMP we don’t need do thermo-cycle, so cooling is not required.
The photosensor sits in the center of the heating element, surrounded by LEDs. On top of this there’s a reasonably complex optical system:
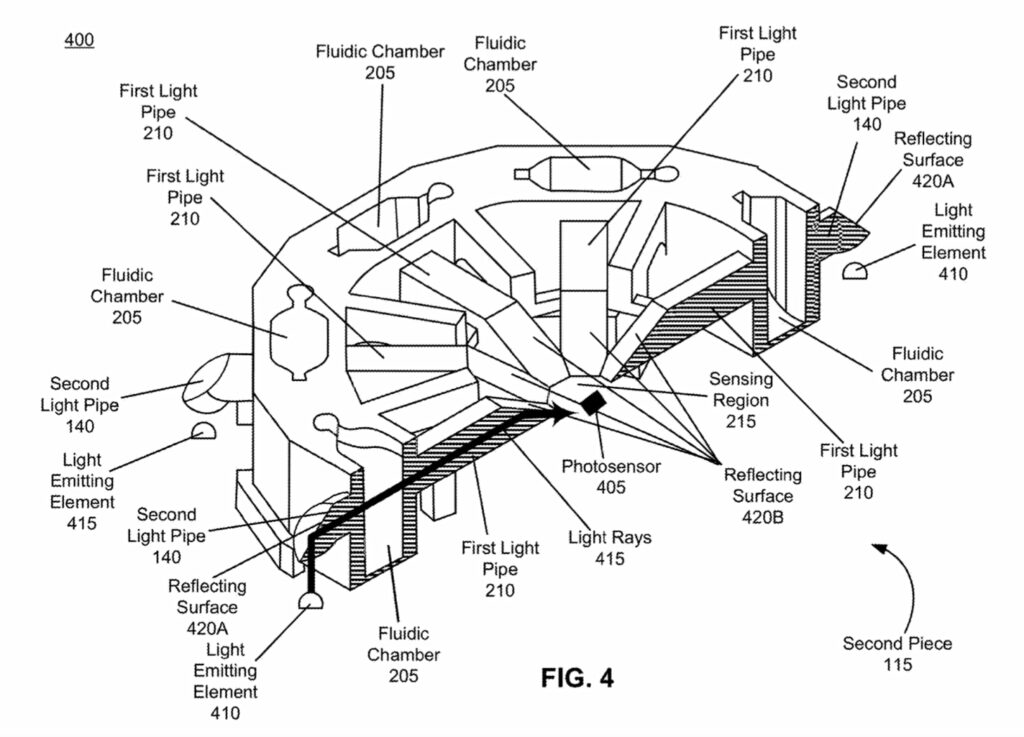
The sample will flow down from the prep tube and be exposed to a reagents ending up in a number of fluidic chambers. This is kind of neat, because as we can see in the image above each LED is paired with a chamber. The light from the LED is reflected through the chamber and down on to a single photosensor.
This allows you to multiplex to some degree, using a single sensor and multiple LEDs which you can switch on/off. Naively you might think that for COVID-19 you only need a single measurement. But the FDA authorization makes it clear that the device also has a “Positive Internal Control (PIC) and Lysis Internal Control (LIC)” which must also show the expected results.
Looking at Brad’s teardown images, we can see that Lucira have only placed 4 of the possible 8 LEDs in the COVID-19 instrument. So Lucira could potentially expand the capability of the device to other targets/variants:

I wasn’t able to identify the photosensor used here, but I suspect it doesn’t cost more than $1, similarly the LEDs likely cost almost nothing. The microcontroller is an STM32F030C6, a simple 32bit ARM microcontroller with 32K of flash, 4K of RAM. This also usually costs ~$1. Overall I suspect the BOM cost is somewhere in the region of $5. The photosensor could potentially be the most expensive part if this turns out to be a high sensitivity photodiode…
The optical system appears to be all plastic, here you can see the light pipes. They show up as different colors, it’s possible there are embedded filters. You can also see the heat conductive paste that is used to couple the heating element to the rest of the unit:

The result of all this is a platform, where much like qPCR, you can monitor the amplification process in order to determine the presence/absence of your target. The patent shows some example experimental data:
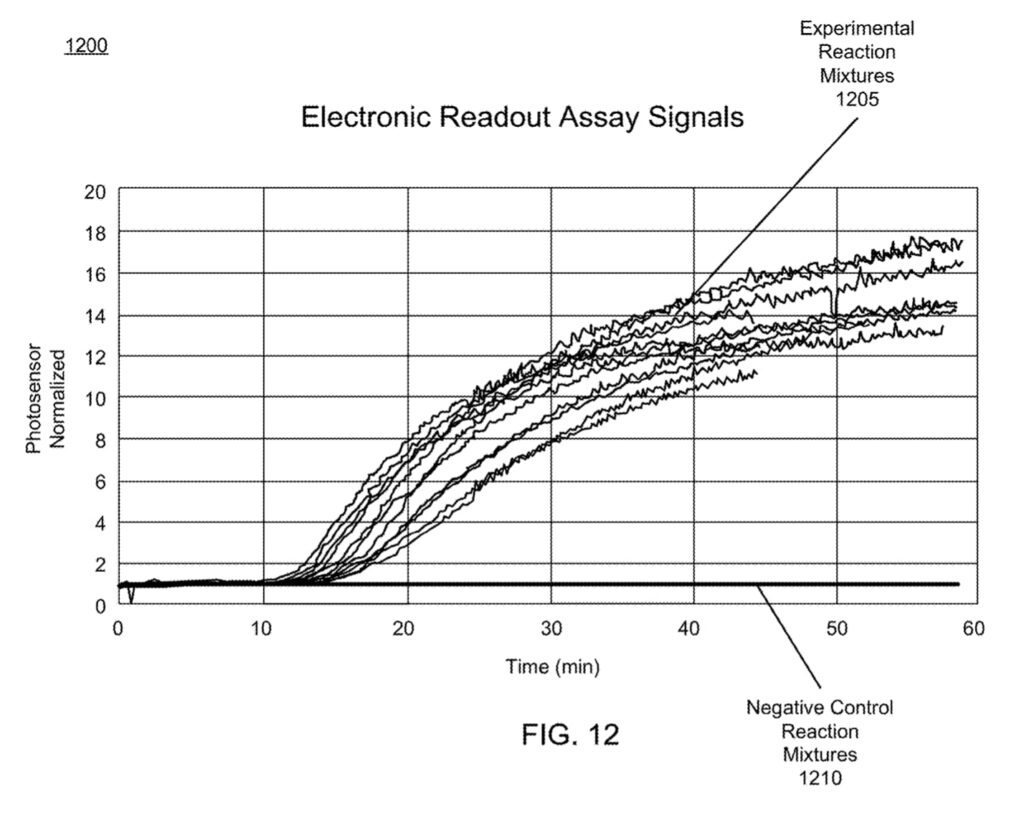
While I suspect the electronics doesn’t cost much, I’m still impressed they can sell this for $55, as it’s a reasonably complex optical/reagent system. I’d be curious to know what their margins are. Overall I find the approach pretty neat. No doubt BARDA (who gave them $21.9M in 2018) must consider this a prescient funding decision.
Lucira IPO’d in February, but it seems that their share price has been rather unstable. It will be interesting to see what happens with Lucira and if they and gain adoption outside of COVID19 diagnostics.