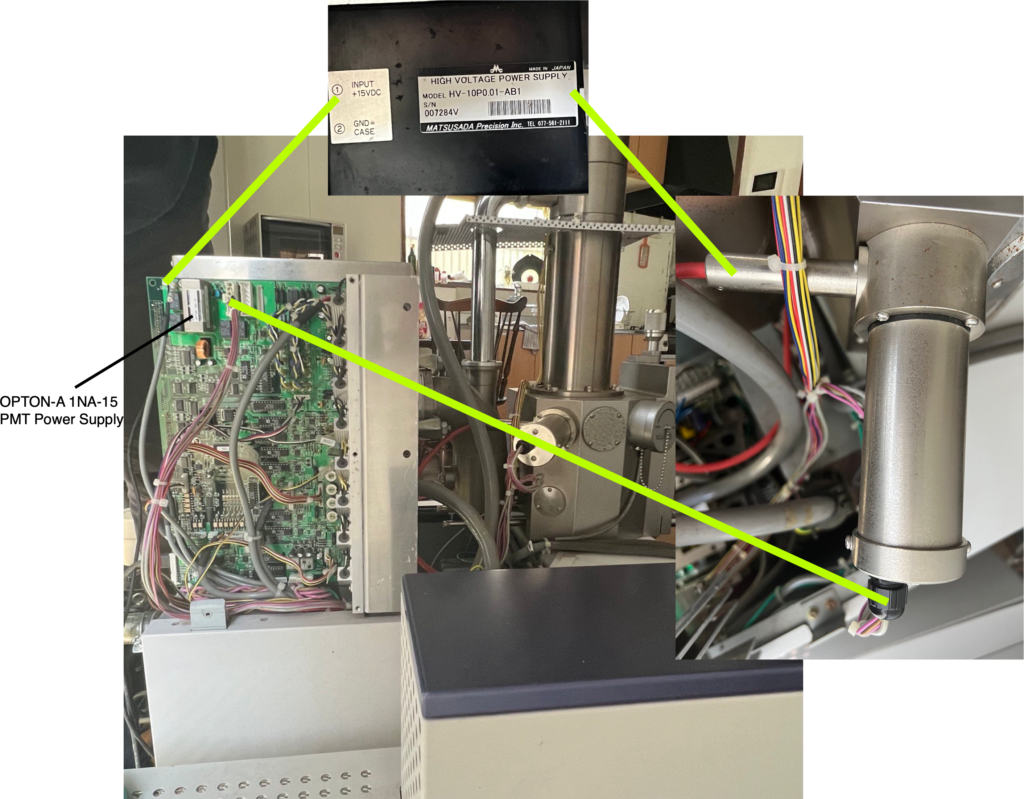
Here’s the modeling code from the substack post.
from math import comb
from math import log10
import sys
def tprb(n, k, t):
totalp = 1
for i in range(0,k,1):
p = 1 - (t / (n - i))
totalp = totalp * p
return 1-totalp
ctoffset = 45
ctslope = -3.36
def predictCT(fragments,ampregion,fragmentsize,genomesize):
totalfragments = genomesize-fragmentsize
ontargetfragments = fragmentsize-ampregion
targetfragments = fragments*(ontargetfragments/totalfragments)
ct = ctoffset+log10(targetfragments)*ctslope
return ct
genomesize = 30000
totalng = 10
qpcrampregion = 90
basesperng = 1771000000000
rrnafraction = 0.6
fragmentsize = 1000
starttargetfraction = 0.000000001
targetfraction = starttargetfraction
totalfragments = (basesperng*totalng)/fragmentsize
reads = 100000
targetfragments = totalfragments*targetfraction
totalfragments_norRNA = ((totalfragments-targetfragments)*(1-rrnafraction)) + targetfragments
# Vary reads count
#print ("total fragments, reads, target fragments, probability of >1 read, probability of >1 read (no rRNA), predicted CT")
#for creads in range(reads,100000000,1000000):
# print (totalfragments, " ", creads, " ", targetfragments, " ", tprb(totalfragments,creads,targetfragments), " ", tprb(totalfragments_norRNA,creads,targetfragments),predictCT(targetfragments,qpcrampregion,fragmentsize,genomesize))
# Vary target fraction
#print ("target fraction, total fragments, reads, target fragments, probability of >1 read, probability of >1 read (no rRNA), predicted CT")
#while targetfraction < 1:
# targetfragments = totalfragments*targetfraction
# print (targetfraction, " ", totalfragments, " ", reads, " ", targetfragments, " ", tprb(totalfragments,reads,targetfragments), " ", tprb(totalfragments_norRNA,reads,targetfragments),predictCT(targetfragments,qpcrampregion,fragmentsize,genomesize))
# targetfraction *= 10
# Vary both
#print(", ", end='')
#while targetfraction < 1:
# targetfragments = totalfragments*targetfraction
# print(targetfraction, ", ", end='')
# targetfraction *= 2
#print()
#for creads in range(reads,5000000,100000):
# print(creads, ", ", end='')
# targetfraction = starttargetfraction
# while targetfraction < 1:
# targetfragments = totalfragments*targetfraction
# print (tprb(totalfragments,creads,targetfragments), ", ", end='')
#
# targetfraction *= 2
# print()
# Vary both - CT
print(", ", end='')
while targetfraction < 1:
targetfragments = totalfragments*targetfraction
print("{:.2f}".format(predictCT(targetfragments,qpcrampregion,fragmentsize,genomesize)), ", ", end='')
targetfraction *= 2
print()
for creads in range(reads,5000000,100000):
print(creads, ", ", end='')
targetfraction = starttargetfraction
while targetfraction < 1:
targetfragments = totalfragments*targetfraction
print (tprb(totalfragments,creads,targetfragments), ", ", end='')
targetfraction *= 2
print()