Bionano Genomics (Bionanomatrix)
It might seem odd for Bionano to be on a list of DNA sequencing companies given that Bionano have not publicly discussed the developing a DNA sequencing platform. Bionano are instead focusing on genome mapping. Mapping technologies give long range information on the structure of the genome (megabases, at >100bp resolution). This allows you to detect large scale structural variation in a genome (such large scale changes are associated with cancer for example). In contrast to this, most current DNA sequencing technologies focus on accurate short range information, providing single base resolution reads of about 100bp.
However mapping and sequencing exist in a continuum. As sequencing read lengths increase (into the mega base range with some approaches) they can more easily address issues of structural variation. Similarly, as the resolution of a mapping technology decreases (ultimately to single base resolution) it turns into a long read sequencing technology.
Because of this technological continuum, I’ve included mapping companies that have their own unique sensing system. And in this post I discuss Bionano Genomics.
Business
Bionano genomics (originally Bionanomatrix) was founded in 2003, to date they have raise 132.1M USD (according to crunchbase) from a number of investors [1], they also received about 3M USD in SBIR grants [3]. Their first commercial instrument launched in 2012. They now appear to be filing for an IPO seeking to raise 34.5M USD [2]. Genomeweb also reported that Bionano received 1.7MUSD in revenue Q1 2018 (net loss 3.8M). On March 31st they had 7.6MUSD in cash. Meaning without further financing they can last a couple for quarters. In 2017 total revenue was 9.5MUSD. So the 2017 average revenue per quarter was a little higher than Q1 2018s. They currently have 65 employees (28 in research).
Technology
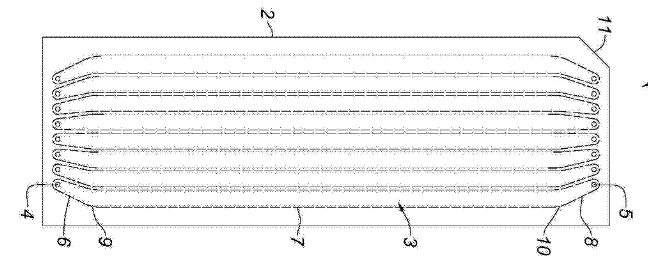
Bionano’s devices were at least partially developed from work undertaken at Lund University and Princeton. Patents refer to a 2004 paper co-authored by Bionano Genomics founder Han Cao [5]. The paper shows 100nm nanochannels:
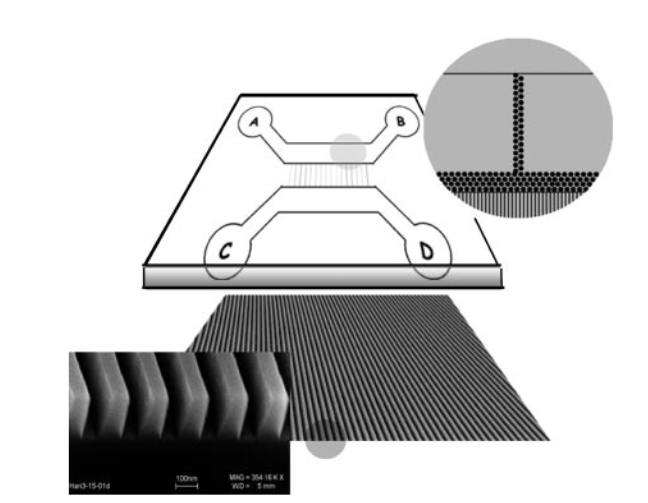
In this paper double stranded DNA is driven through the nanochannel under a bias voltage. There’s a nice image of some stretched DNA:
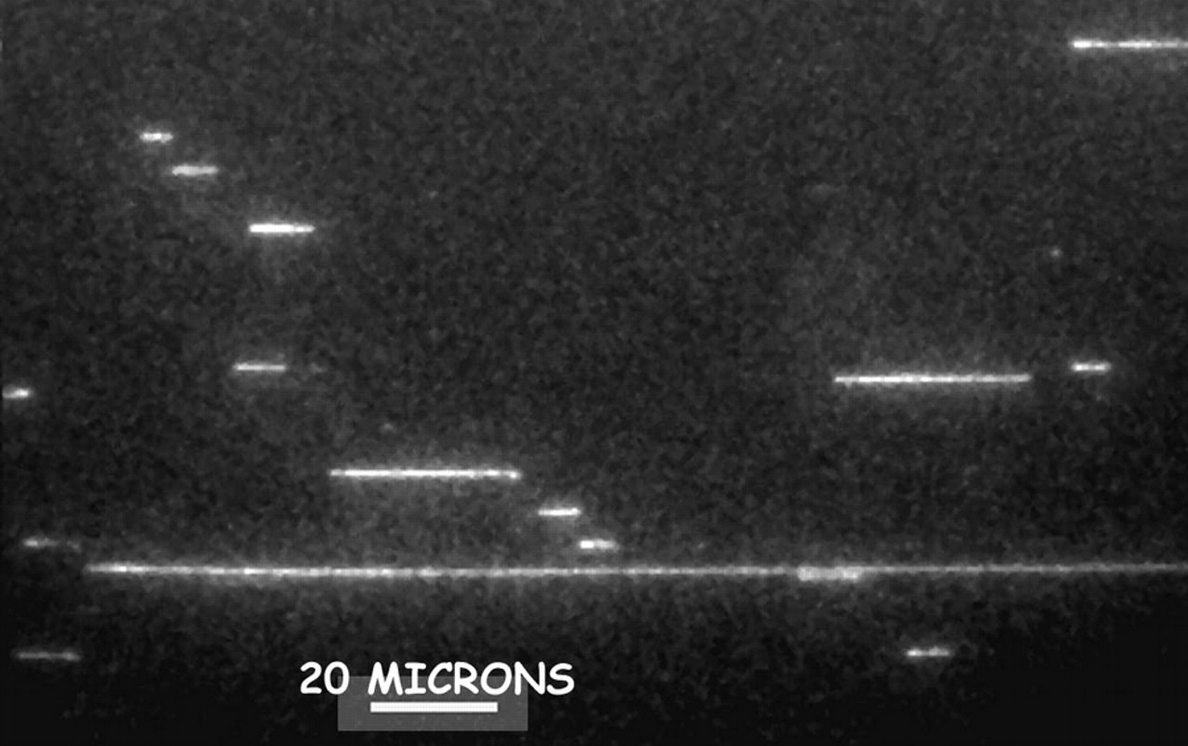
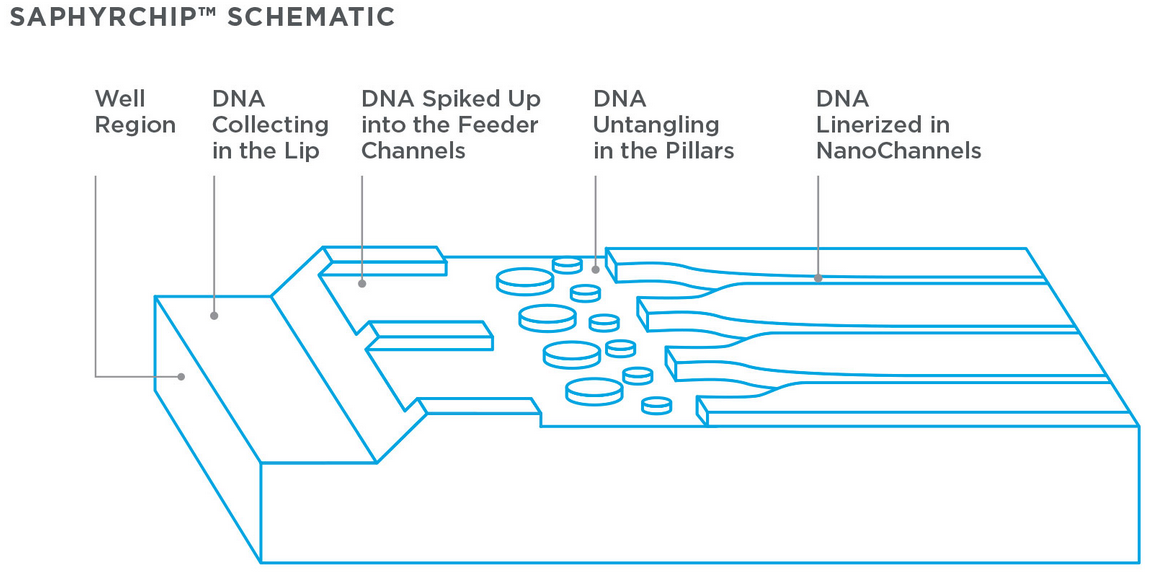
Here is DNA is labelled with TOTO. This is an intercalating dye which labels the entire strand. The Bionano chip seems to be a development of this, and there’s a nice schematic on the Bionano website:
As you can see the chip is somewhat more advanced than that presented in the 2004 paper. In particular pillars have been added to untangle DNA. Once the double stranded DNA is linearized in the channels it can be imaged.
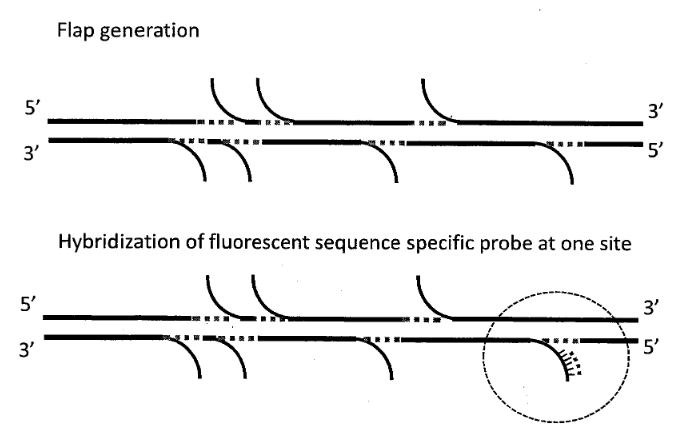
But just imaging stained DNA isn’t very useful. In order to create a structural map of the fragments, we need some kind of specific labelled to create a “map” of the fragment. Most typically this is performed using a nicking endonuclease. These enzymes recognize particular target DNA sequences and create small single stranded “nicks” in the DNA:
Fluorescent probes can then be attached to the nicked locations to allow their locations to be imaged. Using this information you can obtain a map of the DNA strand, telling you the relative locations of the nicking enzyme recognition sites. You can then compare these molecule maps against known references. By looking for large scale rearrangements, it’s possible to determine where structural variation occurs.
The system resolution is likely limited by several factors, and their site states that the Irys system resolution is 1.5 kbp or ~500nm, this would make sense as the optical resolution of a diffraction limited system.
In addition to there currently released platform Bionano do have one patent where they incorporate sequencing into their platform [4], but given this is from 2009 it seems likely that they have not pursued this approach currently. Some more digging might reveal other interesting approaches in their patents.
Notes
[1] According to Crunchbase, investors include: Domain Associates , Novartis Venture Fund , Legend Capital , Battelle Ventures , Gund Investment, LLC , KT Venture Group , Innovation Valley Partners , 21Ventures and Ben Franklin Technology Partners of Southeastern Pennsylvania.
[2] https://www.genomeweb.com/business-news/bionano-genomics-seeks-raise-345m-ipo
“Bionano also revealed that it had $1.7 million in revenue in the first quarter of 2018 and a net loss of $3.8 million, or $1.16 per share. Its R&D expenses for the quarter were $2.4 million and its SG&A expenses were $2.9 million. As of March 31, the company had $7.6 million in cash and cash equivalents.
In 2017, the company had $9.5 million revenues and a net loss of 23.4 million, or $7.66 per share. Its R&D expenses were $12 million in 2017, and its SG&A expenses totaled $14.1 million.
As of December 2017, Bionano had raised approximately $129.3 million through sales of its preferred stock. In addition, in 2016, it entered into a secured term loan facility with Western Alliance Bank, under which it has borrowed $7 million. The loan facility requires it to raise $5 million from the sale of equity securities by Aug. 3 of this year.
Bionano said that as of March 31, it had 65 employees, including 28 in sales, sales support, and marketing, 28 in research and development, four in manufacturing and operations, and five in general and administrative positions. Of its employees, 57 are located in the US and eight elsewhere.”
[3] https://www.sbir.gov/sbc/bionanomatrix-inc
[4] https://patents.google.com/patent/CA2744064A1
[5] http://www.pnas.org/content/pnas/101/30/10979.full.pdf