Iridia (was Dodo Omnidata)
While putting together my list of synthesis companies, one particular stood out. Not least because of its original name, Dodo Omnidata (which is awesome) [3]. But also because the technology is significantly different from anything else on the list being inherently single molecule. The company also seems to be relatively unknown.
For these reasons, I’m writing up some quick notes.
Business
Dodo Omnidata was founded in 2016. They seem to have raised ~400K in seed funding in 2017. An SEC filing shows they raised ~2MUSD this June. Jay Flatley (ex-Illumina CEO) is on the board. The initial 400K came from Tech Coast Angels according to Crunchbase. It’s not clear where the most recent raise came from, but with Jay on the board, it seems possible there’s a connection to Illumina Ventures.
Technology
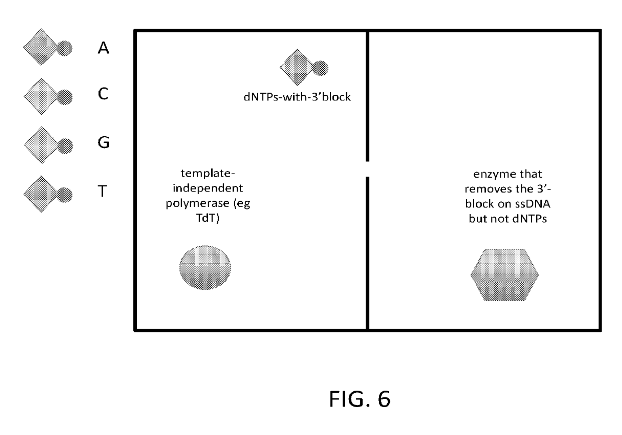
There’s not much on the website, but there is a 134 page patent. I’ve barely skimmed it but what’s clear is that they suggest using nanopores for DNA synthesis:
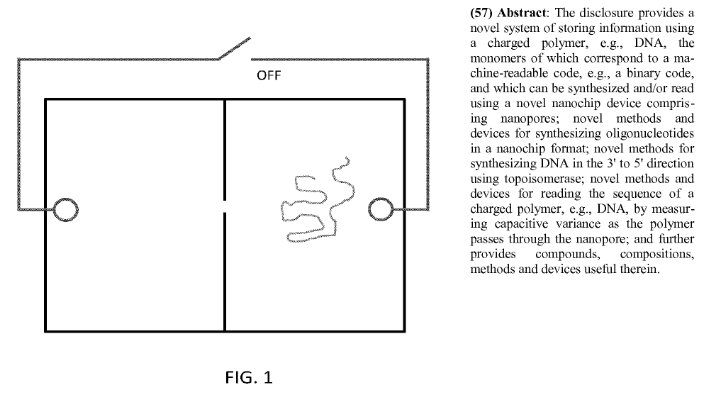 From by quick skim, it appears that what they suggest is driving a strand of DNA through a nanopore with a bias voltage. In this way they can move it between two chambers. In itself I don’t believe that is particularly novel. What’s neat is that because enzymes are too big to go through the nanopore they can selectively expose the strand to different enzymes under electrical control.
From by quick skim, it appears that what they suggest is driving a strand of DNA through a nanopore with a bias voltage. In this way they can move it between two chambers. In itself I don’t believe that is particularly novel. What’s neat is that because enzymes are too big to go through the nanopore they can selectively expose the strand to different enzymes under electrical control.
They use this for synthesis by having one chamber containing a template independent polymerase (a polymerase that just adds any base you give it) and a base with a terminator on it (so only a single base is added). My guess is that you’d flow bases in cyclically. If you want to incorporate a base into the strand, you flip the voltage and pull the strand through the nanopore. Leave it for a while to incorporate the base, then pull it back out.
Back on the other side of the pore, another enzyme comes in and removes the block on the strand. As single nucleotides can also pass through the pore, it’s desirable to have an enzyme that only removes terminators on bases incorporated into the strand.
In practice I would imagine the whole system can be arrayed. And you’d be flowing bases onto one side of an array. How competitive this system is with other enzymatic approaches is something I don’t know. But it seems pretty neat!
Notes
[1] 2018 SEC Filing: https://www.sec.gov/Archives/edgar/data/1708118/000170811818000002/0001708118-18-000002-index.htm
[2] http://www.freepatentsonline.com/WO2017151680A2.pdf
[3] In case you’re curious about the binary encircling the old Dodo Omnidata logo it converts to Data Vida in ASCII. Vida is Spanish for life, and I assume is a reference to the tagline “Data for Life” also on their banner.