As part of my series of posts on DNA sequencing companies I wanted to write up my notes on Stratos Genomics.
Business
Stratos have raised a total of 55.8MUSD according to Crunchbase [0], most recently in January 2018. Investors include Roche Ventures and Fisk Ventures. The Roche investment is particularly interesting, as it’s possible that there is some synergy with their acquisition of Genia. The company was founded in 2007 by Allan Stephan, Mark Kokoris.
Glassdoor reviews [4] are pretty interesting, indicating an intense work culture. Normal work hours of 8am to 7pm with extended work required are also listed on most job postings.
Technology
My notes here are based on their 2008 patent [1] with a few observations from a recent ASHG poster [3].
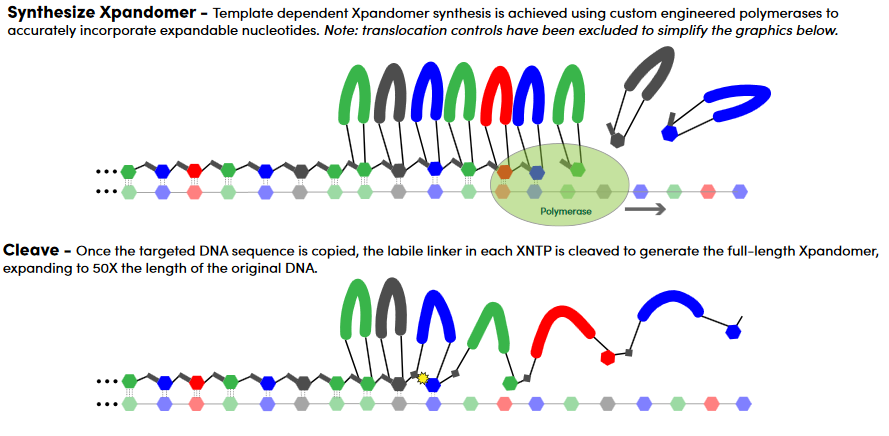
The Stratos approach is termed “Sequencing-by-expansion”. Notionally the idea is to use the original DNA as a template to create a much longer fragment. Each single base in the original strand results in many bases (or other reporter molecules) in the synthesized copy.
For example, the original strand might be: ATG… The synthesized strand derived from this could read: AAAAAAAAA—TTTTTTTTT—GGGGGGGGG.. (-s might indicate non-nucleic spacers).
Such a process could be beneficial in nanopore sequencing. Achieving single base resolution using a nanopore has proved challenging, and error rates are relatively high. Replacing each base with multiple copies, or a different, large, reporter has multiple potential benefits.
Firstly, it gives you more time to observe the signal from each nucleotide. Controlling the speed of translocation is sometime problematic in nanopore systems. By increasing the length of each observation, you potentially increase accuracy, and reduce the need for other approaches to slowing down translocation.
The second benefit is that most of the time only a single base type should be sitting in the pore. In current nanopore platforms signal from multiple base positions are convoluted. De-covoluting the signal to obtain the original sequence is a complex and error prone process. If only a single type of base is sitting in the pore, the signal is vastly cleaner making the original sequence simple to determine.
You could of course, also incorporate other labels into the synthesized strand, potentially marking the recognition process even easier.
The Stratos approach appears to be to perform their expansion process, and then sequence the expanded fragments on their own platform. Patents refer to alpha-hemolysin and it seems likely that they are targeting the use of this process with a protein nanopore platform.
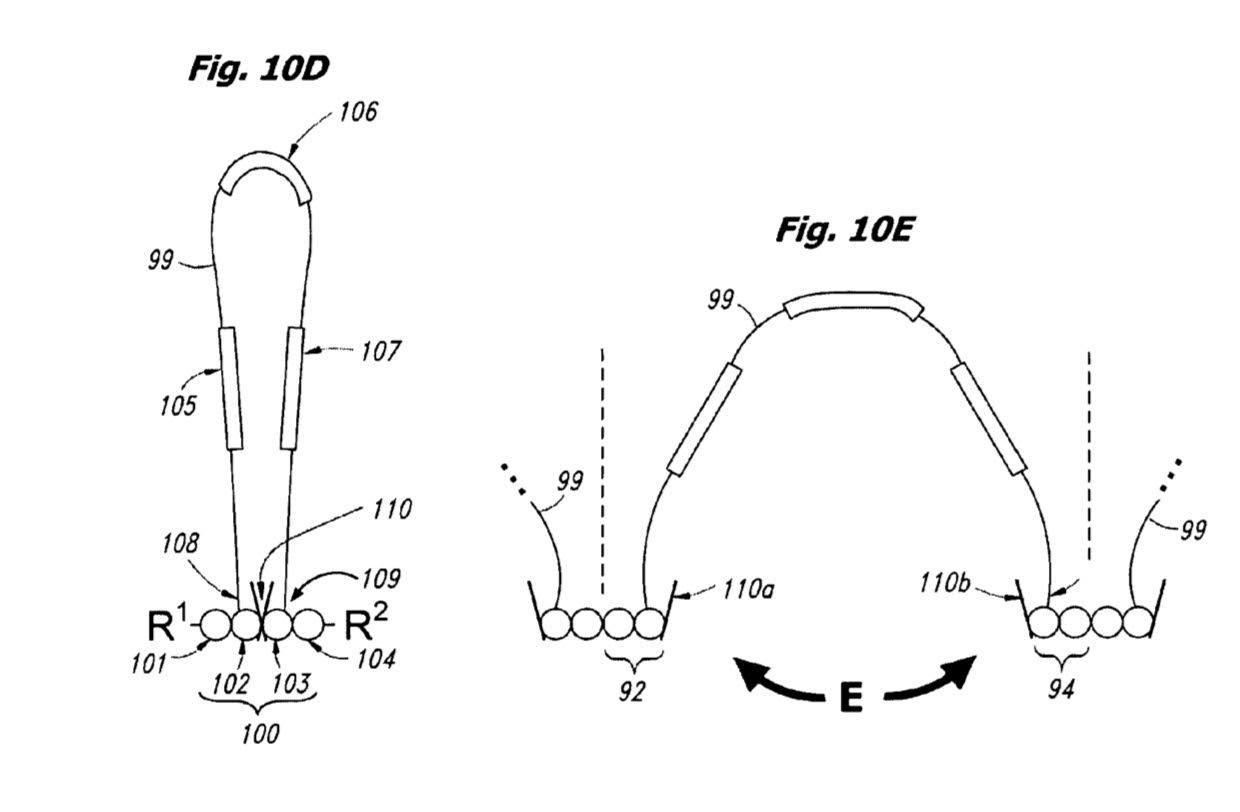
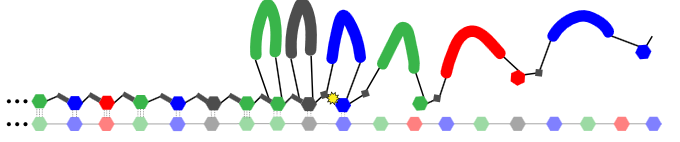
A number of expansion methods are described in the patent (but appear to have changed more recently, see below). The basic process appears to be 2mers with rather long “looped” labels, like those shown below:
The bases are joined using a cleavable linker. To build the “expandomer” these novel oligos are hybridized and ligated to the template. In itself this seems like a potentially problematic step, as I can imagine that hybridization and ligation of 2mers is very specific. Ligases will also need to be selected that work with these complex labels. Perhaps because of this a recent poster shows the use of a polymerase to incorporate these loop labeled nucleotides:
In the polymerase process there seems to be a cleavable intermediate between each base to which adjacent bases bind. It seems likely that using a polymerase to incorporate these bases is less error prone than the ligation based approach in the patent.
Once all these “loop labeled” oligos have been incorporated the synthesized strand is melted away from the template and cleavage of the linker between the parts of the loop is performed.
The result is an “expanded oligo”:
This new sequence, contains spacers/linkers and is not a suitable template for further amplification. However it can be size selected and put through a nanopore for sequencing.
The process seems neat, but I’d bet there’s a lot of work in getting the chemistry working efficiently. The ASHG poster shows experimental results, but these don’t give much indication of the raw read error rate. However they appear to be able to detect single base mutations with some reliability.
Interesting approach, and as experimental results are starting to appear, no doubt one we’ll be hearing about more in the not too distant future.
UPDATE: I’ve been contacted to clarify that: “Sequencing-by-Expansion uses mixed-composition molecules attached to long tethers as base-specific reporters in the newly synthesized strand” and “chemistry has progressed (since the ’08 patent) to using an engineered polymerase that incorporates individual, reporter-tagged, expandable nucleotides in a process similar to PCR.”. There’s a patent covering the polymerase engineering [1a], I’ve not checked if it covered the nucleotides themselves. It would be interesting to read more about these.
Notes
[0] Crunchbase: https://www.crunchbase.com/organization/stratos-genomics
[1] https://patentimages.storage.googleapis.com/12/0f/30/e12fa7aae73ba4/US7939259.pdf
[1a] https://patents.google.com/patent/WO2017087281A1
[3] https://static1.squarespace.com/static/5b1825728ab722032661ec0a/t/5bda53b38a922d1e357a8348/1541034932444/Stratos+Genomics+ASHG18.pdf
[4] Selected glassdoor reviews:
Pros
They have some great ideas for products, some of which are highly innovative.
Cons
Management, specifically Mark Kokoris, is very condescending towards employees, which has resulted in a high turnover due to his mismanagement resulting in many talented employees leaving the organization.
Sadly, cronyism is also a huge problem here. If you fit right in, you’ll advance into management quickly, regardless of whether you have the right skill set. If not, you’ll be blacklisted from promotions.
Advice to Management
Increase employee wages, decrease mandatory long hours, and give those with talent some recognition.
Pros
Interesting/ promising technology for next-gen DNA sequencing. The CSO is a clever scientist. The company has obtained good funding from a major pharma/ diagnostics company, which may eventually buy SG and its technology.
This may be a great place to get some serious (Charles Dickens’s type) real-life experience before a better job.
Cons
Many:
1. A company is run by 1 man, with his iron fist. The man (Mark Kokoris, CSO) is a mega micro-manager, and is very paranoid (although very clever, and has a phenomenal memory). It’s always either his way, or the highway.
2. The company environment is more suited to a 3rd world sweat-shop, than to a USA-based tech company. If you are OK with that, get a job there. Work 11+ hours a day, rarely hear “thank you” for your work, constantly get disciplined, be prepared to be thrown out for anything you may think or say differently from what is accepted by the boss.
3. Environment is paranoid: no privacy — all e-mails can be and are read; anything you say outside of accepted line will be prosecuted; divisive people management style — everything is piece-mailed, management prevents employees from seeing the big picture; when people are let go or leave, they just disappear without a chance to communicate with their coworkers; the reason to fire an employee can be simply a suspicion of “treason” (e.g. being interested in other jobs outside SG) or some kind of disagreement, esp. of work-style nature, with the boss (even though they will pretend there was a legit reason and concoct some text with legit-sounding, but usually ridiculous or untrue content). Leaving the company for another job is considered treason, and the person leaving is labeled a traitor. I don’t know of anyone who left and received a good reference.
4. Benefits suck: small vacation, low pay for the long hours, no overtime. Parking is not covered by the company (and it’s expensive in Belltown). No sick time (you get sick – it comes out of your vacation)
5. All work, no play. It’s different from dedication. This is more of a voluntary concentration camp environment.
6. Obsolete lab equipment (although with new funding, they are starting to buy better things)
7. Marginal lab safety practices (but getting better now).
Advice to Management
it’s hopeless… no advice… No, there are a few:
11+ hours work per day is no guarantee of efficiency. For better efficiency: (1) appreciate your employees, and create a real team-work environment. (2) Instead of piece-mailing and dividing people, promote real (not declared) exchange of ideas — for god’s sake, do overview presentations for all employees, not just project-by-project. Promote flow of ideas between departments. Be more transparent. (3) Allow flexibility of work schedule: you’ll actually get more hours from people — people will not be dead-tired and unmotivated. Tiredness is not productive. (I know you don’t believe me, but this is how it works everywhere else)
(4) Stop spying on people. This is not healthy.
I have been working at Stratos Genomics full-time
Pros
They bump you up in pay fairly quickly.
Full benefits.
Bonus days off in the summer.
An extremely caring, dedicated CEO.
Large, open lab space.
Beautiful view of the Sound.
Stock shares.
They’re willing to train you up.
Cons
The 55 hour work week. If you leave at 7pm you get made fun of because “You don’t leave when you hear the 7pm alarm bell.” It’s not necessary for “real science” either. Work weeks end up being 60-65 hours.
The amount of managers who talk behind people’s backs. And it’s the directors and VPs who do it.
The pay is mediocre and down right terrible if you figure out your hourly rate. Depending on your position, they don’t even pay you for certain hours.
The benefits are also mediocre. Only $200 for glasses or contacts and only every other year.
Management acts as if the world is ending whenever any little mistake is made. Their response to fixing a mistake is always a major overreaction, too.
Advice to Management
Stop talking behind people’s backs and no more 55 hour work week.
I worked at Stratos Genomics full-time (More than 3 years)
Pros
Stratos Genomics has a lot of smart, dedicated scientists working there doing interesting, fast-paced work. It’s a good place to learn new skills and learn from others, especially if you’re just starting out and have a lot of energy to devote to it for a year or two.
Cons
The managers at Stratos Genomics foster a toxic culture. Their goal is to get as much work out of people as they can, pressuring people to work longer hours, take fewer breaks, and work more weekends than other biotech companies in the area do. On top of the long hours, they also gossip about their employees and ascribe the worst motivations onto others as they can. Stratos Genomics has very high turnover, and whenever someone leaves the company, the managers smear their reputation among the remaining employees. It’s a really unpleasant working environment.Show Less
Advice to Management
Good people have options on where they work, and unless you want to only hire inexperienced, recent college graduates, you need to significantly change the way you do things. Your reputation in Seattle is very poor among scientists working at other biotech companies and unless you change your ways by, for instance, hiring a management firm to advise you, I can’t imagine that you will be able to correct your bad working culture.