The Lunatic from Unchained Labs
Unchained Labs
I originally came across the Lunatic when investigating Quanterix’s flow cells. Quanterix flow cells are manufactured by ex-Sony DVD processing unit Stratec. Stratec also manufacture flow cells for the Lunatic, a UV-Vis spectrophotometer made by Unchained Labs. And the name alone was enough to make me want to investigate further. Unchained was recently acquired for $435 million by Carlyle. The company expects to generate $75M in 2021 and has 170 employees.
Unchained have a large portfolio of instruments. None of which I’ve ever heard of, it’s fascinating to me that companies like this can find relatively healthy niches while I assume having a relatively small market share.
Unchained has a storied history, having acquired Trinean, Freeslate, and AVIA Biosystems. They have a portfolio of products covering UV-Vis, scattering, Raman based particle identification, and lab automation. I get the general impression that their focus is on drug development, which might explain why I’ve not heard of them…
In this post, I wanted to delve into “The Lunatic” a little. This is a UV-Vis spectrophotometer which they position against the Nanodrop.
A Brief History of UV-Vis for Nucleic Acid Quantification
The Lunatic is a UV-Vis spectrophotometer and appears to be marketed against the Nanodrop as a tool for detecting impurities in nucleic acids. The basic UV-Vis method looks at nucleic acid absorbance at 260nm and compares this to compares this to the absorbance from proteins and other contaminates peaking at 280nm.
260nm is obviously way down in the UV range which means you need a UV capable spectrophotometer. This implies a UV light source, and UV compatible optics (fused silica) making the instrument more complex, and expensive.
Typically these A260/A280 measurements would be taken using a UV-Vis spectrophotometer such as the Beckman DU530 (which I’ve been tearing apart on my blog). These instruments use light sources covering the visible and UV range. A grating splits this out and a slit is used to select a small range of wavelengths. The grating therefore has to physically move so as to direct a single part of the spectrum toward the slit.
The instrument will scan across wavelength passing the monochromatic light through the sample and measuring absorbance. This is detected with a single sensing element (a photodiode):
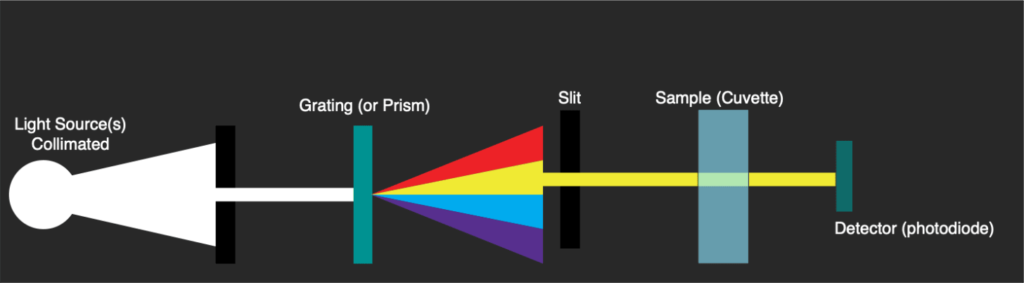
Scans take some time, because a stepper has to physically move the grating to select a wavelength. The photodiode then registers the amount of light absorbed one wavelength at a time. These instruments have a few issues, scans can take more than a minute, and you generally need at least 100ul of material.
The Nanodrop provides a solution to these issues, requiring only 1ul of material, and giving a readout in a few seconds. It does this by using a different architecture:
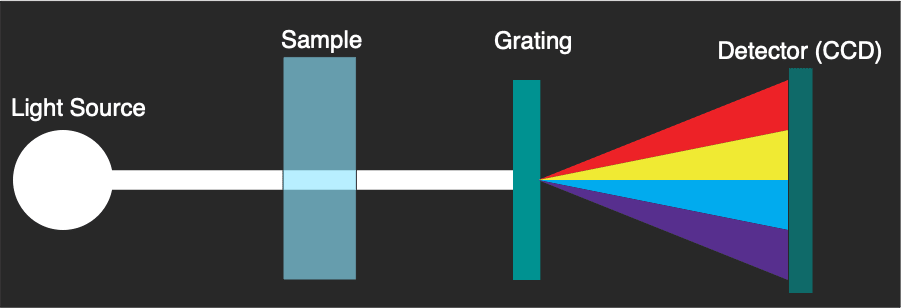
Essentially spread spectrum light is sent through the sample, some is absorbed and the remaining light comes out the other side. Only then is a grating used to split the light and the absorbance spectrum measured. The spectra is registered on a linear image sensor (CCD, or potentially a CMOS sensor in newer instruments). No moving parts are required, and measurement is near instantaneous.
Rather than using a cuvette (sample container) on the Nanodrop the sample is directly sandwiched between two fiber optic cables, which allows for very low sample volume:

For general purpose UV-Vis applications, this approach likely has a number of disadvantages, but for nucleic acid quantification it appears to work just fine.
The Nanodrop doesn’t require any consumables, and from what I understand is a pretty robust instrument, rarely requiring servicing.
The Lunatic
The Lunatic sets itself up as an alternative to the Nanodrop. This is a microfluidic chip based system which performs a similar function to the Nanodrop, and similarly is marketed toward nucleic acid quantification:
There are different chips available for the Lunatic, but they all essentially draw the solution into a detection region. As far as I can tell there’s no active fluidics in this system, it’s all driven by capillary action:
Unchained labs suggest that the Lunatic has similar sensitivity and reproducibility to the NanoDrop. But that the Nanodrop shows significant variation between instruments:
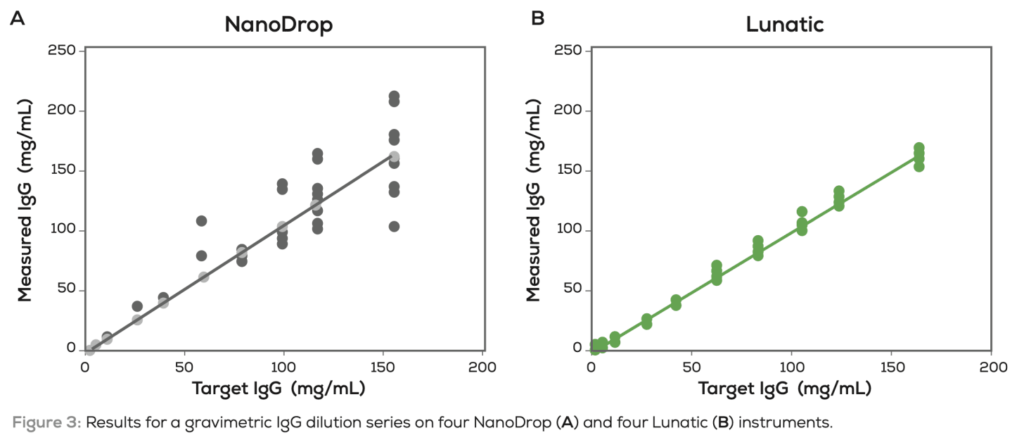
In some ways this feels like a calibration issue, in that the Nanodrops are showing the necessary reproducibility, but that they are not typically calibrated to the same extent as the Lunatic. It’s also not clear to me that the Nanodrop is the right instrument to compare the Lunatic with, perhaps Denovix who position themselves as a higher sensitivity alternative might make a more interesting comparison. The Lunatic does appear to have a higher dynamic range than the Nanodrop, and this maybe useful in some scenarios.
The major disadvantage of the Lunatic of course is that you need to buy chips to use it. From what I can tell, this works out at about $4 per sample. The chip likely as some advantage in terms of ease of use and clean up, but overall I find it hard to see a strong advantage over the Nanodrop and other platforms.
But with this instrument being marketed toward drug discovery, perhaps even a small edge over the Nanodrop coupled with a healthy portfolio of instruments, is enough.