Random Notes
Collection of notes I wanted to put somewhere… nothing to see here.
Translocation and dwell times:
| Translocation time through nanopore (dwell time) | https://41j.com/blog/wp-content/uploads/2023/07/image.png |
| Single molecule diffusion on hard, soft and fluid surfacesRhodamine 6G (R6G) and Rhodamine 123 (R123), both of which can readily absorb on the three selected model surfaces by hydrophobic interaction.octadecyl-triethoxylsilane (OTE) | https://41j.com/blog/wp-content/uploads/2023/07/image-1.png |
| Imaging of single molecule diffusion.phospholipids carrying one rhodamine dye molecule | https://41j.com/blog/wp-content/uploads/2023/07/image-2.png |
| Direct Measurement of Single-Molecule Diffusion and Photodecomposition in Free SolutionIndividual rhodamine- 6G (R6G) and rhodamine-labeled 30-base single-stranded DNA (ssDNA) (DNA- R6GThe predicted full-width diffusion distances, for R6G and DNA-R6G at a 100-ms exposure time, are 75 pixels and 35 pixels (each pixel 200nm so 15 and 7micron). | https://41j.com/blog/wp-content/uploads/2023/07/image-3.png |
| Comparison of different approaches to single-molecule imaging of enhanced enzyme diffusion | https://arxiv.org/abs/2012.15424 |
| A single molecule visualization of DNA diffusion and partitioning in model porous materials. | https://scholarworks.umass.edu/cgi/viewcontent.cgi?article=2071&context=dissertations_1 |
| Picoquant | Diffusion coefficient in water at 25°C (298.15 K) in 10−6 cm2s-1 for Atto655-NHS: 4.25 ± 0.06Which works out as 8.94427 microns per 100ms frame? |
DNA stretched on surfaces:
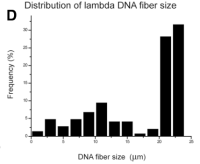
DNA molecules (48.5 kb) on test surfaces, yielding a factor of 2 kb/um [1].
Papers
[1] Dynamic Molecular Combing: Stretching the Whole Human Genome for High-Resolution Studies
Early paper, good reference on stretching length. Used 300um/s stretching speed.
[2] Molecular Combing of Single DNA Molecules on the 10 Megabase Scale
Describes how to stretch DNA without breaking it, 900um/s stretching speed. Nice details on stretching rig.
[3] Image processing for optical mapping
Algorithmic notes on optical mapping algorithms.
[4] A simple and optimized method of producing silanized surfaces for FISH and replication mapping on combed DNA fibers
Size distribution of Lambda DNA (shown above)
Home Isolation experiments (kids):
Videos:

At home
http://www.biotech.iastate.edu/publications/lab_protocols/DNA_Extraction_Onion.html
TEACHER PREPARATIONEdit
- Set up hot water bath at 55-60� C and an ice water bath.
- For each onion, make a solution consisting of one tablespoon (10 ml) of liquid dishwashing detergent or shampoo and one level 1/4 teaspoon (1.5 g) of table salt. Put in a 1-cup measuring cup (250 ml beaker). Add distilled water to make a final volume of 100 ml. Dissolve the salt by stirring slowly to avoid foaming.
- Coarsely chop one large onion with a knife and put into a 4-cup measuring cup (1000 ml). For best results, do not chop the onion too finely. The size of the pieces should be like those used in making spaghetti. It is better to have the pieces too large than too small.
- Cover chopped onion with the 100 ml of solution from step 2.
The detergent dissolves the fatty molecules that hold the cell membranes together, which releases the DNA into the solution. The detergent, combined with the heat treatment used in step 5, causes lipids (fatty molecules) and proteins to precipitate out of the solution, leaving the DNA. The salt enables the DNA strands to come together.
- Put the measuring cup in a hot water bath at 55-60� C for 10-12 minutes. During this time, press the chopped onion mixture against the side of the measuring cup with the back of the spoon. Do not keep the mixture in the hot water bath for more than 15 minutes because the DNA will begin to break down.
- Cool the mixture in an ice water bath for 5 minutes. During this time, press the chopped onion mixture against the side of the measuring cup with the back of the spoon. This step slows the breakdown of DNA.
- Filter the mixture through a #6 coffee filter or four layers of cheese cloth placed in a strainer over a 4-cup measuring cup. When pouring the mixture into the strainer, avoid letting foam get into the measuring cup. It can take more than an hour to recover most of the liquid. The filtering can be done in a refrigerator overnight.
- Dispense the onion solution into test tubes, one for each student. The test tube should contain about 1 teaspoon of solution or be about 1/3 full, whichever is less. For most uniform results among test tubes, stir the solution frequently when dispensing it into the tubes. The solution can be stored in a refrigerator for about a day before it is poured into the test tubes. When the solution is removed from the refrigerator, it should be gently mixed before the test tubes are filled.
STUDENT INSTRUCTIONSEdit
The process of extracting DNA from a cell is the first step for many laboratory procedures in biotechnology. The scientist must be able to separate DNA from the unwanted substances of the cell gently enough so that the DNA is not broken up.
Your teacher has already prepared a solution for you, made of onion treated with salt, distilled water and dishwashing detergent or shampoo. An onion is used because it has a low starch content, which allows the DNA to be seen clearly. The salt shields the negative phosphate ends of DNA, which allows the ends to come closer so the DNA can precipitate out of a cold alcohol solution. The detergent causes the cell membrane to break down by dissolving the lipids and proteins of the cell and disrupting the bonds that hold the cell membrane together. The detergent then forms complexes with these lipids and proteins, causing them to precipitate out of solution.
PROCEDURE
- Add cold alcohol to the test tube to create an alcohol layer on top of about 1 cm. For best results, the alcohol should be as cold as possible. The alcohol can be added to the solution in at least three ways. (a) Fill a pasteur pipette with alcohol, put it to bottom of the test tube, and release the alcohol. (b) Put about 1 cm of alcohol into the bottom of a test tube and add the onion solution. (c) Slowly pour the alcohol down the inside of the test tube with a pasteur pipette or medicine dropper. DNA is not soluble in alcohol. When alcohol is added to the mixture, all the components of the mixture, except for DNA, stay in solution while the DNA precipitates out into the alcohol layer.
- Let the solution sit for 2-3 minutes without disturbing it. It is important not to shake the test tube. You can watch the white DNA precipitate out into the alcohol layer. When good results are obtained, there will be enough DNA to spool on to a glass rod, a pasteur pipette that has been heated at the tip to form a hook, or similar device. DNA has the appearance of white mucus.
Fluoresence optical signatures of plastics
https://www.en.uni-muenchen.de/news/newsarchiv/2014/langhals_plastikmuell.html
Gloves, which gloves to use?
Which gloves to use:
http://amo-csd.lbl.gov/downloads/Chemical%20Resistance%20of%20Gloves.pdf
Nitrile gloves:
Acetone – fair
Ethanol – excellent
Isobutyl – alcohol excellent
Isopropyl – alcohol excellent
Methanol – fair
Methylene Chloride – fair (listed as incidental contact, and double glove)
Latex gloves:
Acetone – good
Ethanol – excellent
Isobutyl – alcohol poor
Isopropyl – alcohol excellent
Methanol – fair
PVC gloves:
Acetone – poor
Ethanol – excellent
Isopropyl alcohol – good
Methanol – good
Viton gloves & Butyl gloves:
Acetone good
Methylene Chloride – (listed as extended contact, Viton. Also recommends Polyvinyl acetate)
inkjet printers!
Protein microdeposition using a conventional ink-jet printer: https://www.ncbi.nlm.nih.gov/pubmed/10723562
HP Journal on Piezo Inkjet printing: https://www.hpl.hp.com/hpjournal/pdfs/IssuePDFs/1988-08.pdf
Agilent patent on inkjet alignment, bio apps: https://patents.google.com/patent/EP1321190A2
Microfab patent on general process: https://patents.google.com/patent/US5658802
Negative back pressure,Venturi throat: https://patents.google.com/patent/US6242266B1/en
Agilent thermal: https://patents.google.com/patent/US6221653B1/en
“a pressure source which provides a gas load pressure valve to the reservoir chamber which is sufficiently negative such that fluid adjacent and facing the orifice is loaded into the head by being drawn into the reservoir chamber through the orifice and delivery chamber, while simultaneously being insufficient to result in ambient atmosphere entering the delivery chamber through the orifice once the head has been loaded and no further fluid is facing and adjacent the orifice. “: https://patents.google.com/patent/US6323043B1/en
Agilent thermal 2: https://patents.google.com/patent/US6458583B1/en
Agilent thermal 3: https://patents.google.com/patent/US6461812B2/en
Using imaging/overlaying to QC inkjet printed microarrays: https://patents.google.com/patent/US6587579B1/en
Agilent use fujifilm heads: https://www.fujifilmusa.com/press/news/display_news?newsID=880165
Tube cleaning with magnetic beads: https://patents.google.com/patent/US6537817B1/en
GMS 417 Arrayer looks like this!

Flowcells:
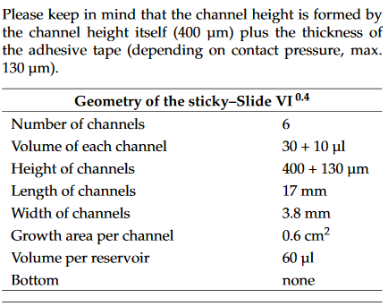
https://ibidi.com/sticky-slides/63-sticky-slide-i-luer.html#/33-pcs_box-15/67-channel_height-01_mm
Holos InterOP:
Overview: https://www.youtube.com/watch?v=VCdn0VgRjkQ
User interface: https://www.youtube.com/watch?v=lHaMYd7J3CY
Glassdoor review:
“I never saw about 10% of my paycheck. Turned out that the person working in HR was taking it for herself. The finance department apparently knew that this was going on. I can only imagine the other types of fraudulent things going on in this company amongst others. The management obviously don’t care about the employees at all, especially after the recent mass layoff. Apparently only around 5 people are left after the last mass layoff, taking whatever funds are left for themselves before they retire or if they ever sell whatever’s left of this sad excuse of a company. Outlook for that isn’t so great though, since the market for cataract surgery devices is so small and is already being outcompeted by larger companies with better talent and technologies like ORA. Honestly, their investors are just sinking dead cash into a lost cause at this point. I hope they finally pull the plug.”
Their old office appears to be up for rent: https://42floors.com/us/ca/pleasanton/5775-w-las-positas-blvd
Sequential Wavefront Aberometer
Website: http://claritymsi.com/ (dead) “Please come back later.” June 2018

Inheco TEC Control 96



Wired for 2 wire RTD 100Ohm Platinium.
Switched DC Open collector output. Outputs 1 and 2 connected.
http://www.schneordesign.com/Avi/WatlowMod/watlow.htm
Output 1 seems to control heating.
Output 2 seems to control cooling.
The Watlow LSTW appears to combine these two singles, and contains an H bridge to control the TEC…
Instrument was limited to 55C. Datasheet suggests 70C max. No cooling below 4C…
I’ve tried the device up to 90C
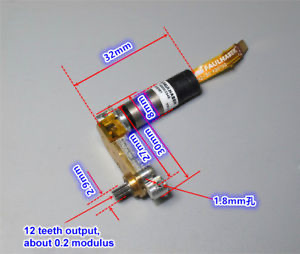
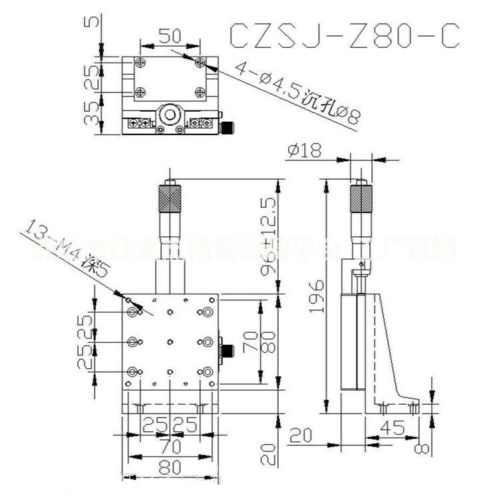
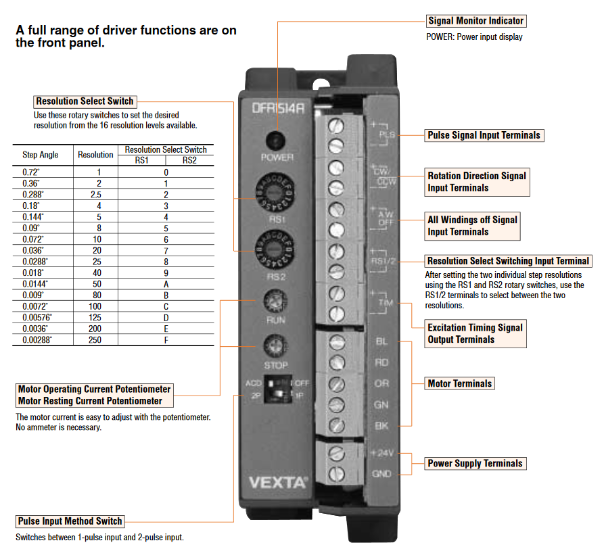
Motor:
miRNASeq:
| Paper | Exosomal | Kits Used | Notes (Sequencer/Tools) | Total Reads | Total Mapped | Mapped miRNA reads | Genomic Reads | Other RNA | Other Unknown Reads | Sequence data available? |
| Analysis of the RNA content of the exosomes derived from blood serum and urine and its potential as biomarkers | Yes | PGM318, not selected for miRNA. | 5 to 6M | 90 to 98% | 2 to 45% | <2% | ~55 to 90% | 2 to 10% | ||
| Characterization and deep sequencing analysis of exosomal and non-exosomal miRNA in human urine | Yes | RNA isolation kits: miRNeasy (Qiagen), miRNeasy with RNeasy MinElute Cleanup Kit (Qiagen), mirVana PARIS (Ambion), Trizol LS reagent (Life Technologies) with mirVana (Ambion), miRCURY (Exiqon), and the Urine Exosome RNA Isolation kit (Norgen Biotek). | Urine Exosomal and Non-exosomal miRNAPGM318 | 7,075,206, average of 786,134 per sample. | 90%+? | 35% | 61% mapped to regions between genes. | <1% ncRNA3% other | ||
| Downregulation of exosomal miR-192-5p and miR-204-5p in subjects with nonclassic apparent mineralocorticoid excess | Yes | TruSeq 1 Small-RNA Library Prep Kit | Illumina NextSeqmiRDeep2 | Yeshttps://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE138556 | ||||||
| A comparison of RNA extraction and sequencing protocols for detection of small RNAs in plasma | RNA extraction: MagnaZol (Bioo Scientific), miRNeasy (QIAGEN)Library prep: CleanTag (TriLink), NEXTflex (Bioo Scientific) and QIAseq (QIAGEN) | Illumina NextSeqCLCBio | 5.8M to 12.8M | 9.5% to 18.9% | Yeshttps://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE118125 | |||||
| Urinary exosome miR‐30c‐5p as a biomarker of clear cell renal cell carcinoma that inhibits progression by targeting HSPA5 | Yes | TRIzol Plus RNA Purification Kit | Illumina HiSeq 2000 | 15.8M | 76% | 62% | 9.5% piRNA | 28% | ||
| Urine microRNA Profiling Displays miR-125a Dysregulation in Children with Fragile X Syndrome | NO – probably not useful | Extraction: Norgen Biotek TaqMan MicroRNA Reverse Transcription Kit | Illumina HiSeq 2500CLCBio | 15.5M to 25.9M | Yeshttps://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE143347 | |||||
| Comparability of the small RNA secretome across human biofluids concomitantly collected from healthy adults | NEBNext small RNA sample library preparation kit | Illumina HiSeq 1000 STAR | 16.6M to 48.2M (40.4M average) | 10% | 10% | miRNA comprised 92.9% and 93.3% of uniquely aligned small ncRNA sequences | Yeshttps://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE122621` | |||
| A combination of circulating miRNAs for the early detection of ovarian cancer | No | QIAzol and the miRNeasy Mini KitTruSeq Small RNA Sample Prep Kit (Illumina) | Illumina HiSeqCOBWeB/StrandNGS | 11.8M to 21M | Passing filter 48% | Adapter containation <5%Smaller than 10nt 4% to 37% |
| Optimizing exosomal RNA isolation for RNA-Seq analyses of archival sera specimens | Yes | Yes | 20 ng | RNeasy Mini Kit TRIzol LSAllPrep DNA/RNA Mini kit | STAR | ~10M per sample | >90% | ~0.5%>500K | Yes | |||
| Combination of Four Serum Exosomal MiRNAs as Novel Diagnostic Biomarkers for Early-Stage Gastric Cancer | Yes | No | 2 ng RNA2 nM seq. | ExoQuick – System BiosciencesMultiplex Small RNA Library Prep Set for Illumina – NEB | ~10M per sample | 73% | Yes | |||||
| Methods for Extracting and Characterizing RNA from Urine: for downstream PCR and RNAseq Analysis | No | No | ignored concentration estimates used maximum volume | Qiazol ReagentmiRNeasy Mini Kit (Qiagen)Miscript II kit (Qiagen)TruSeq Small RNA kit | Hiseq2000miRDeep | 1.6 to 50M | “few percent to more than 25%” | Yes | ||||
| Performance assessment of total RNA sequencing of human biofluids and extracellular vesicles | No | Yes | loading concentration of 1.1 or 1.2 pM | miRNeasyUltra-Clean Production (UCP) columns (Qiagen)HL-dsDNaseTotal RNA-Seq Kit v2 – Pico Input Mammalian (Takara, 634413) | NextSeq 500 | ~15M reads | ~>2% | 90% Protein coding | Yes | |||
| Total Extracellular Small RNA Profiles from Plasma, Saliva, and Urine of Healthy Subjects | No | No | TruSeq small RNA | cutadaptsRNABench | 11.6 million reads | 2%Discarded urine and saliva samples with <0.5% miRNA mapped reads | 68% | 8% rRNA | ~18% too short | No | ||
| Cell and Microvesicle Urine microRNA Deep Sequencing Profiles from Healthy Individuals: Observations with Potential Impact on Biomarker Studies | Both | No? | e.g. 2.8M | e.g. 200K 7%Average is 24.9% | Yes |
Blu-Ray/CD/DVD Optics
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6066758/#ref63
BW DPSS
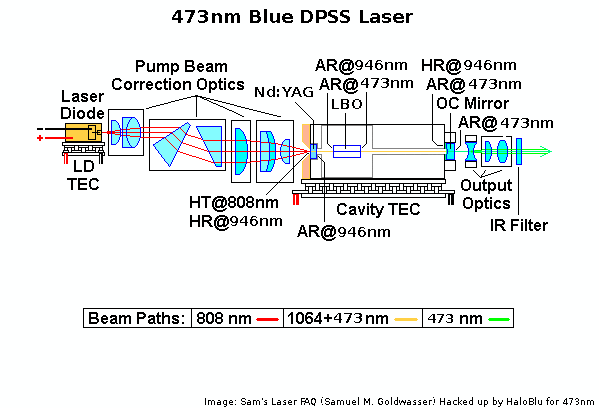
http://repairfaq.cis.upenn.edu/sam/laserscl.htm#sclbwt1
https://sites.google.com/a/dtaviation.com/lasers/dpss
Camera GC1600CH
GigE Vision, Sony ICX274 EXview CCD sensor, auto iris, 25 fpsEdit
Prosilica GC1600H is a 2.0 Megapixel camera with a GigE Vision compliant Gigabit Ethernet interface and Hirose port. Prosilica GC1600H is offered in both monochrome and color models. This camera incorporates the high quality Type 1/1.8 (8.923 mm diagonal) Sony ICX274 CCD sensor with Super HAD CCD technology that provides superior image quality, excellent sensitivity, and low noise. At full resolution, this camera has a frame rate of 25 frames per second. With a smaller region of interest higher frame rates are possible. By default monochrome models ship with no optical filter and color models ship with an IRC30 IR cut filter.
Benefits and features:
- Monochrome (GC1600H) and color (GC1600CH) models
- GigE Vision interface
- Screw mount RJ45 Ethernet connector for secure operation in industrial environments
- Supports cable lengths up to 100 meters (CAT-5e or CAT-6)
- Popular C-Mount lens mount
- Easy camera mounting via standard M3 threads or optional tripod adapterr
- Easy software integration with Allied Vision’s Vimba SDK and compatibility to the most popular third party image-processing libraries.
Options:
- CS-Mount
- Optical filters (IR cut filter/Protection glass)
See the Modular Concept for lens mount and optical filter options.
CoolSNAP CF Camera
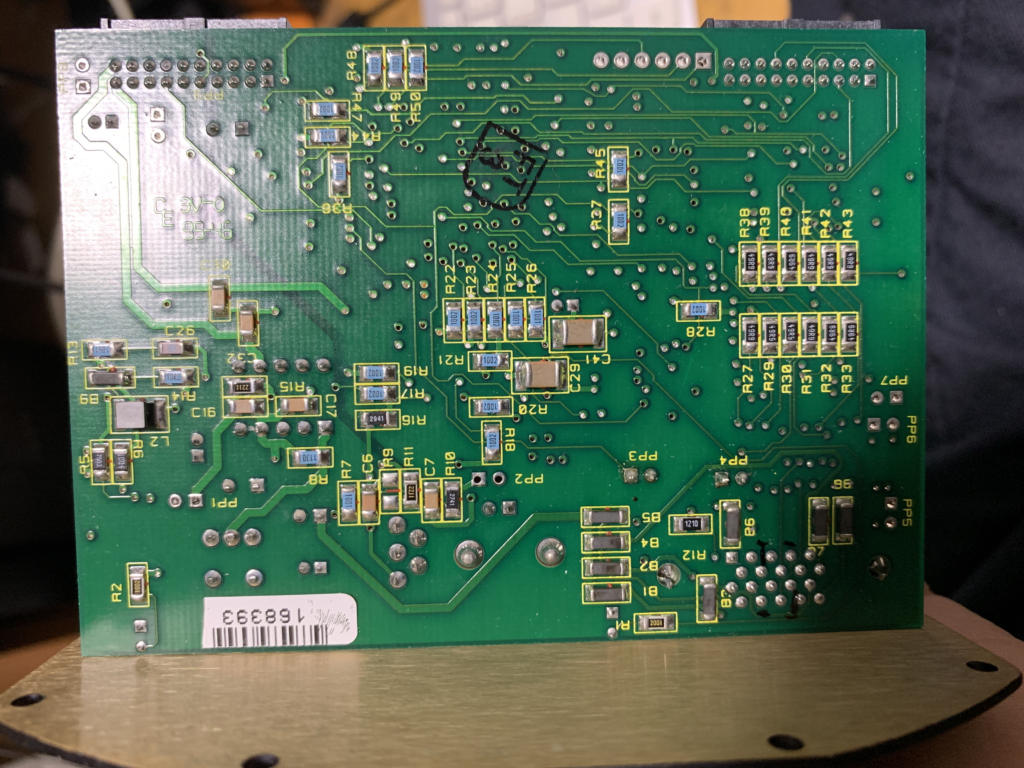

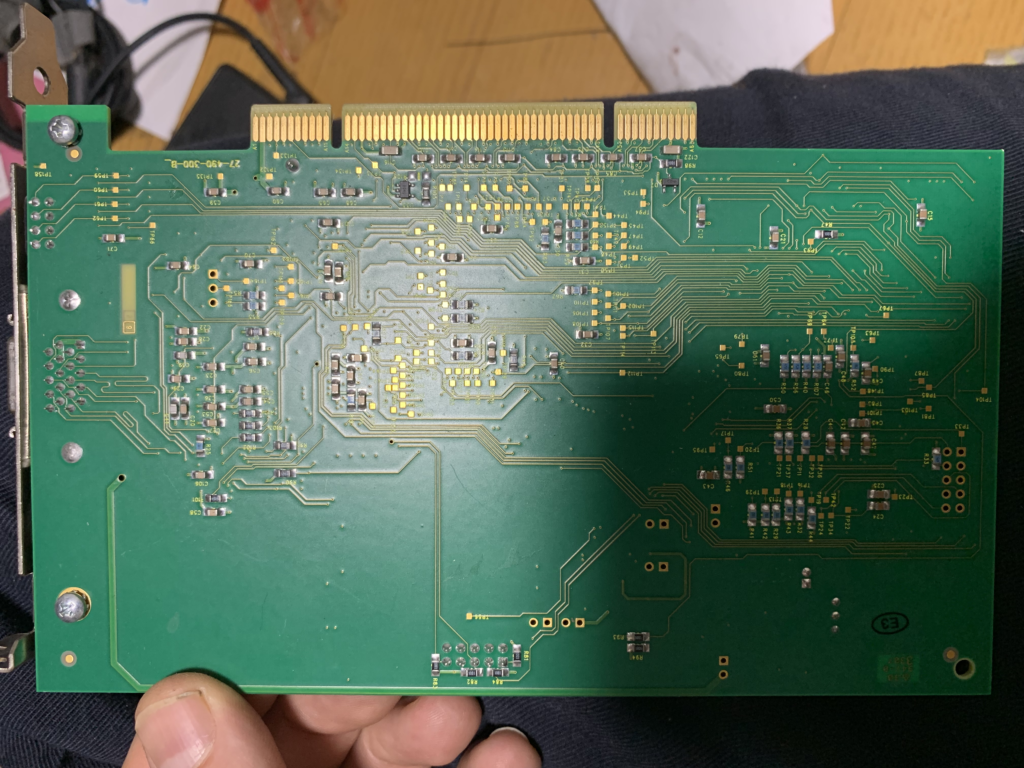
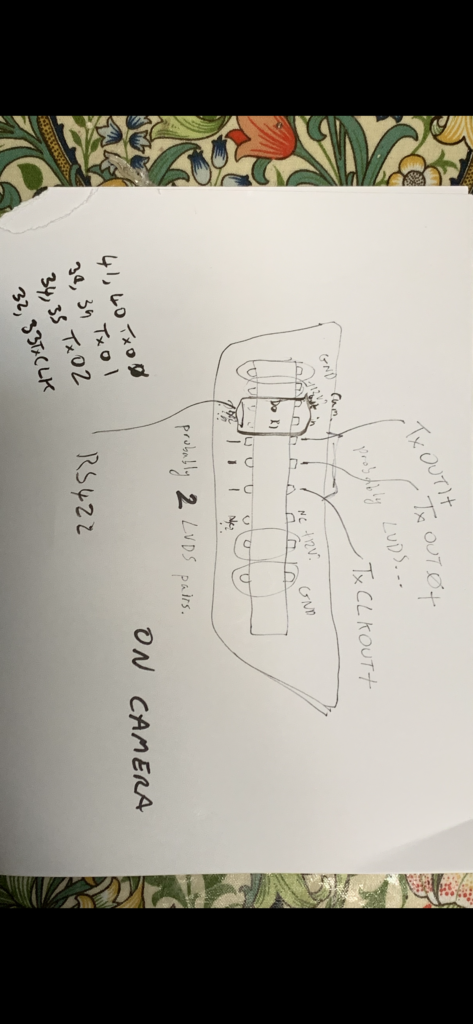
DFR1507A
DLPDLCR2000EVM
Recommended AC adapter:
- TE20A0503F01
LEDs:
- OSRAM red, green, and blue LED – LE BA Q6WM and LCG H9RM
- Red/Amber LED data sheet: https://media.digikey.com/pdf/Data%20Sheets/Osram%20PDFs/LE_BA_Q6WM_v3.2_2015-03-03.pdf
Attempted:
Replace with UV LED: https://www.digikey.com/en/products/detail/inolux/IN-C33BTNU2/9681216
WARNING: PINOUTS DO NOT MATCH, REQUIRES BODGE.
Second option:
Third option (used):
Temp sensor? Measures as 90K
eBay Microscope module
CMOS sensor has marking: HBCFF 02RZ.
Have pixel images (can estimate pixel size).
48 pins, 12 per side.
Board markings: 014101-129-25 26/16 Rev. 01
Best match appears to be MT9P031 or MT9P series.
Responds to I2C commands using this pinout as reference, board supplied with 5v, using 3.3v external pullups. Module contains required 2.8v and 1.8v regulators. One other unidentified regulator? Appears to use parallel bus.
I2C responds on address 0x49. This is not the correct device ID for MT9P. Commands do not appear to match MT9P:
e.g. device ID:
I2C>[0x92 0
NACK
I2C START BIT
WRITE: 0x92 ACK
WRITE: 0x00 ACK
I2C>[0x93 r r
I2C START BIT
WRITE: 0x93 ACK
READ: 0x17
READ: ACK 0xB0
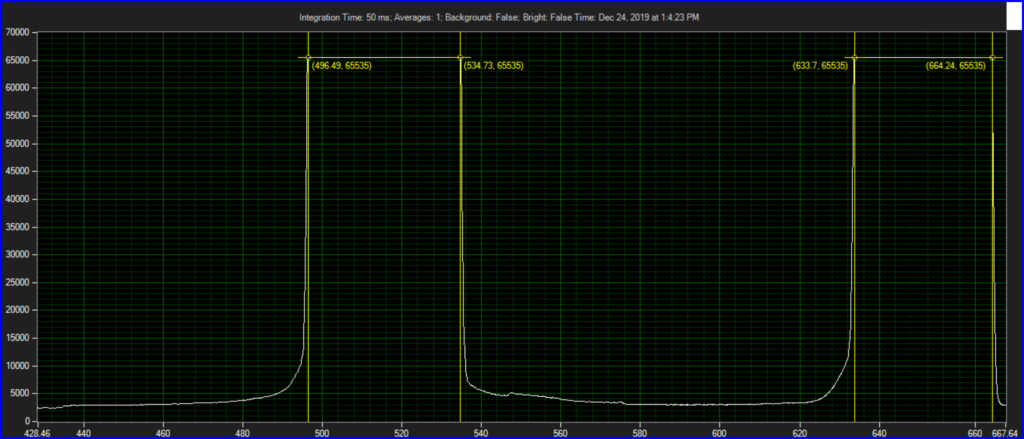
CCD has filters which pass ~550 to 620.
Spectrum of filter on CCD, from Xenon light source:
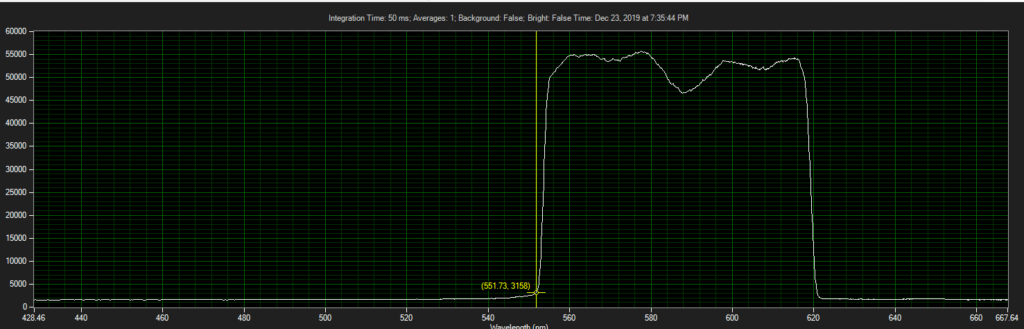
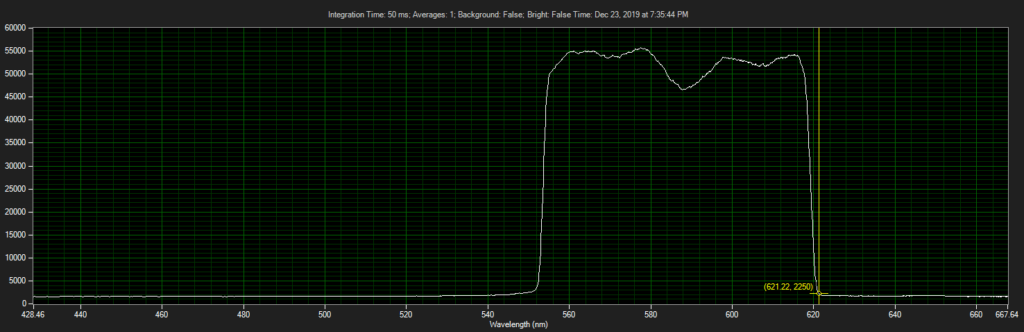
Filter on CCD optics, nearer objective:
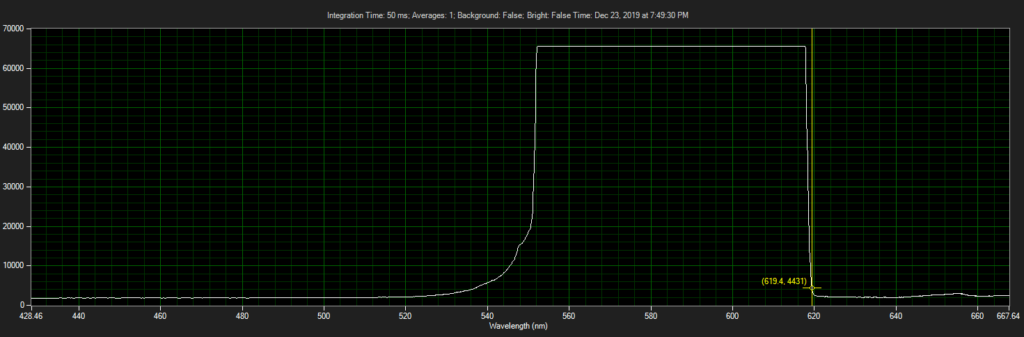
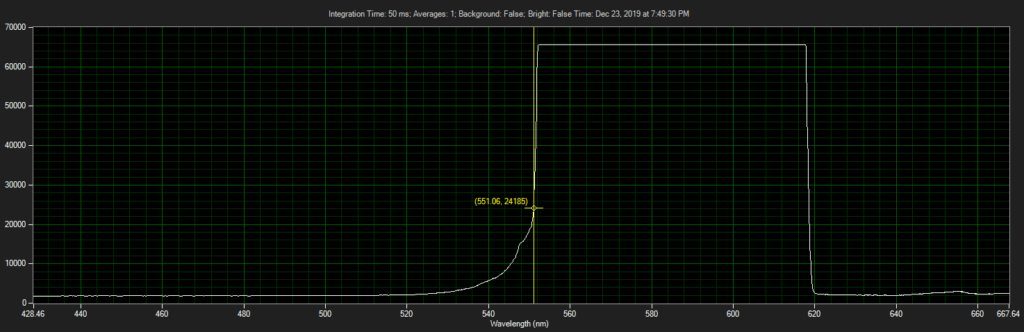
Mirror on objective. Called filter 3 on ebay, reflect toward CCD of Xenon:
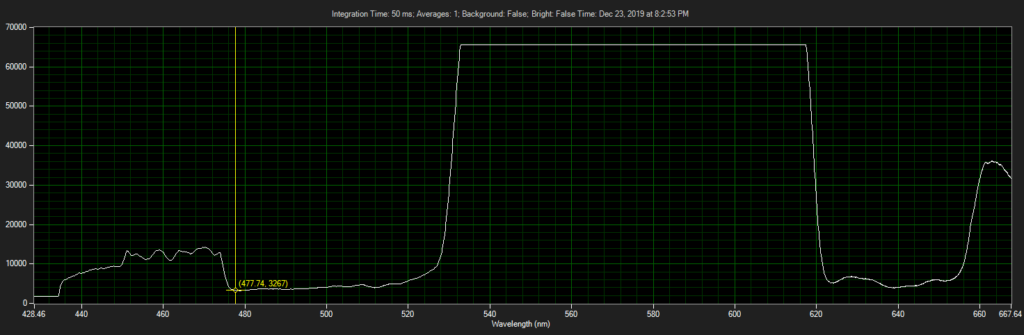
Filter 3, as filter:
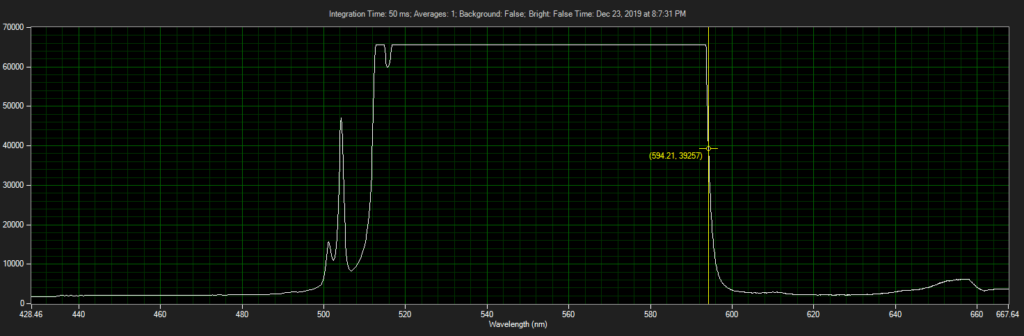
Mirror on laser:
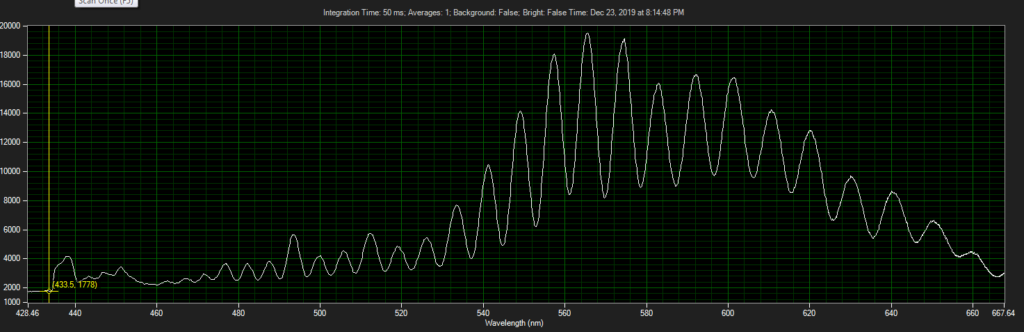
Filter below both LEDs:
Epson DX7


Epson PX-049A etc. printhead

https://dspace.mit.edu/bitstream/handle/1721.1/85435/870681346-MIT.pdf?sequence=2_1
