Singular Genomic Systems S-1 Review
I’ve previously written about Singular Genomics. It now looks like their preparing for an IPO and have filed an S-1. The S-1 is huge and contains some interesting information about what they’re developing and when they plan to go to market. It’s fairly readable and I recommend taking a look if that kind of thing interests you.
In this post I’m going to try and quickly review the technological aspects of the Singular Genomics approach based on the S-1. This document doesn’t really go into specifics (and I’ll refer to other posts on this) but we can get a sense of where they lie technologically in respect to Illumina. So I’m going to dive into the technology and briefly review some of the business aspects at the end of the post. There are two aspects to the Singular Genomics play. The first is the basic sequencing instrument. The second are various sample prep and other analytical approaches they have in development, some of which complement sequencing.
Sequencing
In summary, they look to be developing a 4 lane, (4 color?), Miseq-like instrument. There’s a figure buried in the middle of the document that describes the basic chemistry:
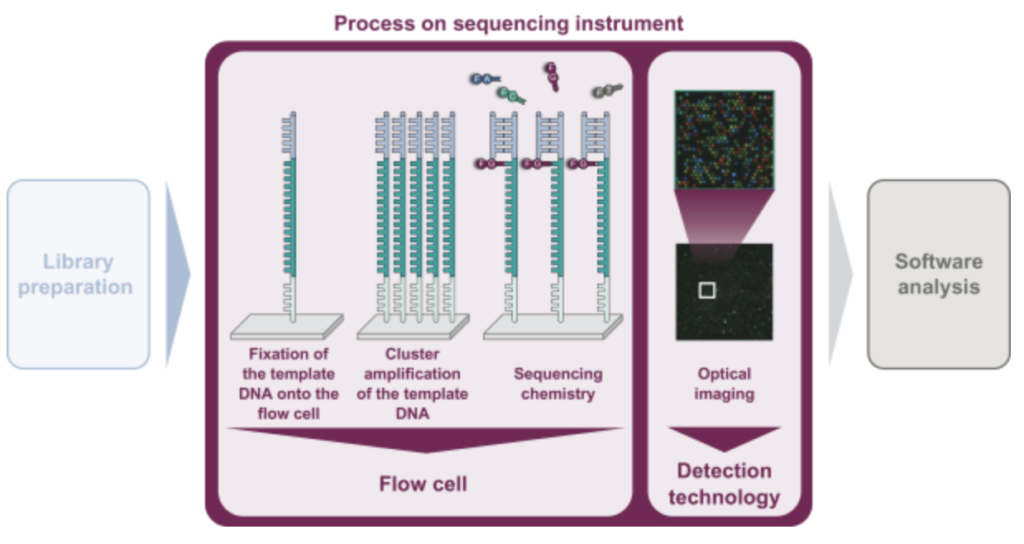
As in Illumina style sequencing, they perform on surface amplification of a single template to generate clusters, and then sequencing-by-synthesis to read out the sequence. The S-1 doesn’t state exactly how they are generating clusters. A quick patent search doesn’t yield anything either. Illumina use a bridge amplification approach, the initial IP was created by Manteia (acquired by Solexa, and then Illumina). I’ve discussed that IP elsewhere, and it appears to have expired. So my guess would be that they’re using this expired IP for cluster amplification.
They don’t appear to be using patterned flowcells, or ExAmp. This makes the approach more like that used on the Miseq (and original Genome Analyzer) than that used on the NovaSeq (and NextSeq 2000). This will limit the density of reads on the flowcell somewhat. From the image above it also looks like a 4 color chemistry, which complicates the optical system somewhat as compared to Illumina’s current generation instruments.
The S1 lends some weight to the idea that they are targeting “MiSeq-like” throughput: “We purposely designed our G4 Integrated Solution to target specific applications and to be capable of competing with other instruments across a range of throughput levels, particularly in the medium throughput segment.”. Flowcells, reagents cartridges, and instruments look fairly familiar:

The projected run times appear to be similar to a Miseq giving a “sequencing time of approximately 16 hours to complete a 2×150 base run.”. Similarly, Illumina quote a 17h run time for their Miseq nano kits at 2x150bp.
As I understand it the Illumina reversible terminator chemistry isn’t yet off patent. The S1 states that they “anticipate initiating an early access program followed by a commercial launch of the G4 Integrated Solution by the end of 2021, with intentions for units to ship in the first half of 2022”. Which suggests that they wont be using Illumina-style reversible terminators unless they believe that IP wont hold up anyway (this seems risky as Illumina are currently using it to block MGI from selling instruments in the US).
The S1 also states that “We in-licensed certain patents and other intellectual property rights from The Trustees of Columbia University” so it’s likely that they’re using Jingyue Ju’s nucleotides, as previously discussed.
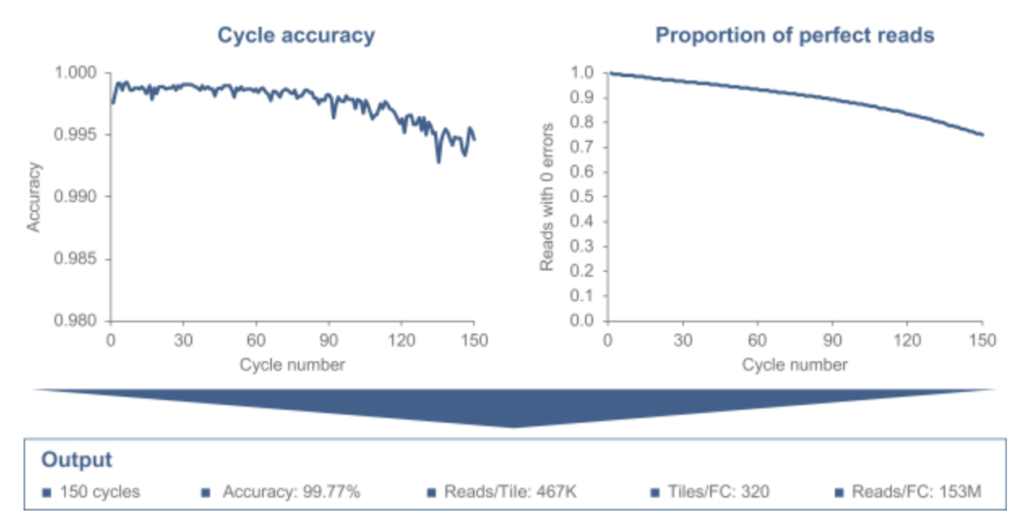
They show some results on sequencing, overall data quality looks like it’s in the same ballpark as current Illumina systems. I’d suggest, much like MGI, it’s a reasonable drop in replacement for Illumina:
Increased data quality though sample prep
Another aspect of the Singular approach appears to be what they call “HD-Seq”. HD-Seq is designed to enable lower error rate reads that can “achieve accuracy levels of Q50” (error rates of 1 in 100,000). It appears to read out both strands of a double stranded fragment and combine the basecalls from both. They pitch this as being particular important for cancer diagnostics through the sequencing of cfDNA. And show some results:
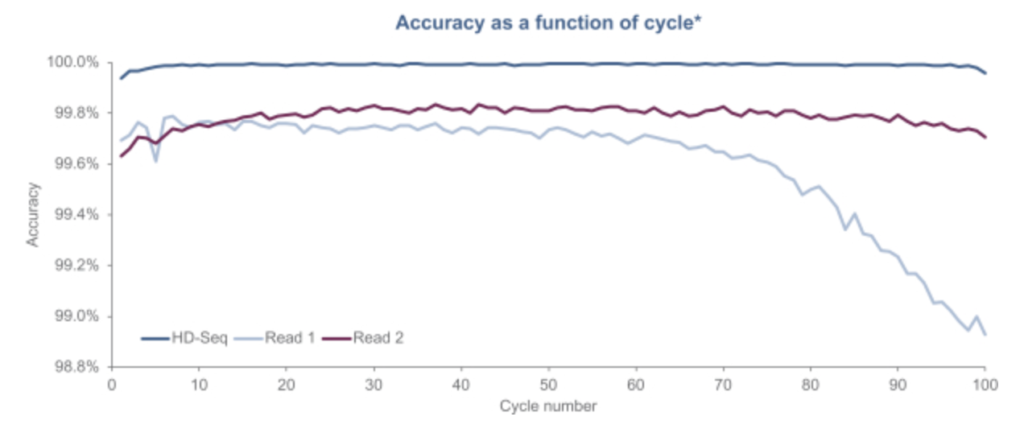
They don’t state how this works exactly, but we can make some guesses. One way of doing this that’s been suggested is to tag the forward and reverse strand with the same index. You can then combine this information, giving you two observations of the same base, reducing the error rate. There’s a patent on this which I guess they may have licensed (though it’s not mentioned in the S1). Other approaches such as the 2D sequencing approaches used by Oxford Nanopore introduce a hairpin, allowing you to read through the forward, and then reverse strand.
You can also use a read pooling/indexing approach (such as 10X use to generate synthetic long reads) to make it easier to pair forward and reverse strands.
Beyond this, I could imagine a neat approach that takes advantage of cluster generation technology. Essentially, flow in double stranded DNA and weakly immobilize it on the flowcell. Then melt it such that the strands separate and attaches to nearby probes/primers on the flowcell. These two templates are then physically close on the flowcell. During image processing/basecalling you can then see that these two templates have a similar sequence (in reverse complement) and likely come from the same source double stranded DNA.
Singular state that their method can “provide higher accuracy than standard single-strand NGS sequencing methods (including ours)” so it’s likely agnostic of sequencing approach. Which suggests to me it’s likely an indexing or hairpin based technology.
Their motivation seems to be “oncology where there is an increasing need for higher sensitivity technology such as rare variant detection in liquid biopsy”. I’m not entirely convinced by this, beyond sensitivity, base modifications seem to be becoming increasingly important for early stage cancer detection. Is the difference between a single base accuracy of Q30 and Q50 critical? Or to put it another way, would you swap two Q30 reads for one Q50? Would be interesting to see more of a justification (or a reference) on the requirement for high quality reads.
Beyond this, they mask out potentially error’d bases: “the base call was only made if there was agreement in the base calls on the complementary strands”. So, while the overall error rate might be lower they limit their ability to detect individual SNPs… using this approach do you still retain the benefits of a lower overall error rate?
If a “read it twice” approach really is of critical importance to cancer diagnostics. There are a number of other techniques that would also work on Illumina’s platform (as discussed above). So this doesn’t feel like a key advantage of the Singular platform.
Other Stuff
Singular seem to be suggesting that they’ve developed a number of other analytical approaches and techniques and have a general purpose “multiomics” platform. The exact methods are not described. But they seem to be pushing further downstream into various applications. Many of which seem to be in 10Xs general territory:
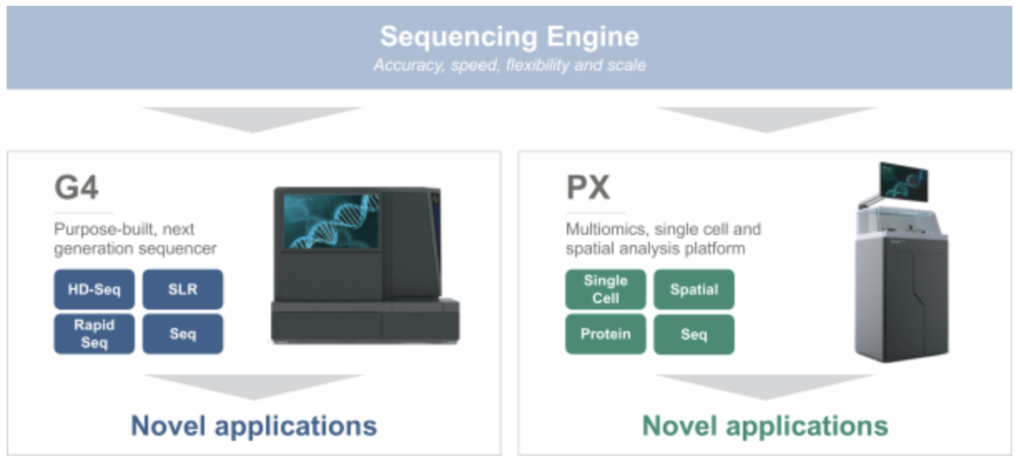
Of particular note is their work on Single Cell and Spatial applications. They also mention Protein expression…
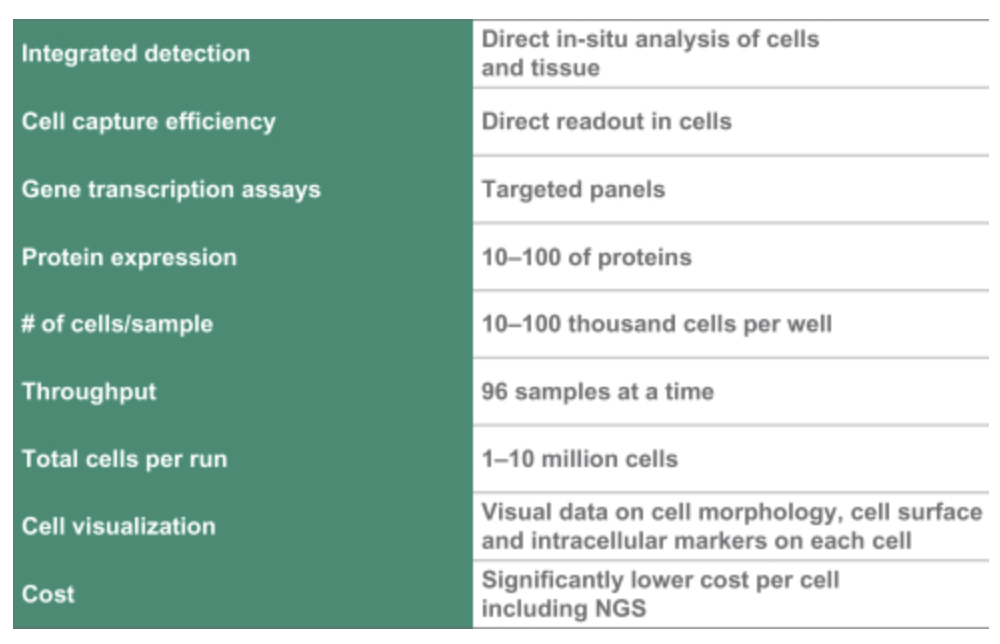
This is worth noting, but there not much in the S1 on the approaches used, and a quick search didn’t pull up any interesting patents here.
Business Stuff
That wraps it up for the technology. The S1 lists current investors, which contains many of the usual suspects:
Entities affiliated with Deerfield Private Design Fund IV L.P, Axon Ventures X, LLC, Entities affiliated with Section 32 Fund 2, LP, LC Healthcare Fund I, L.P, Revelation Alpine, LLC, Deerfield Private, Design Fund I.V., Domain Partners IX, LC Healthcare Fund I, ARCH Venture Fund IX, L.P., Axon Ventures X, LLC.
And that they’ve applied for the NASDAQ symbol “OMIC” which is a pretty cool symbol! They also state that they have 138 full-time employees (106 in R&D).
They state that: “Single cell, spatial analysis and proteomics markets: We are building our PX Integrated Solution to address the single cell and spatial analysis markets, which we estimate to be approximately $17 billion in 2021 based on available market data”. Given that 10Xs 2020 revenue was $298.8M. There’s probably something I don’t get here… where’s the rest of the single cell market?
——
So… those are my initial thoughts on the Singular S1. The basic sequencing approach looks very “Illumina-like” to me. There are a number of other plays like this around (e.g. MGI), and I suspect they will continue to put pressure on Illumina to reduce their consumable prices (which appear to be sold at ~10X cost of goods). But otherwise, I don’t see the approach as opening up new markets or giving us the ability to solve novel research problems.
DISCLAIMER: I own shares in various sequencing companies based on my previous employment. I consult in sequencing, and sequencing applications. And I’m working on a seed stage sequencing related project currently (get in touch if you might be interested in investing in such a project).